S279 point mutations in Candida albicans Sterol 14-α demethylase (CYP51) reduce in vitro inhibition by fluconazole
- PMID: 22252802
- PMCID: PMC3318376
- DOI: 10.1128/AAC.05389-11
S279 point mutations in Candida albicans Sterol 14-α demethylase (CYP51) reduce in vitro inhibition by fluconazole
Abstract
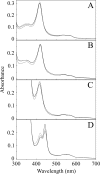
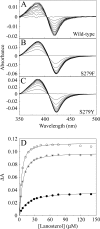
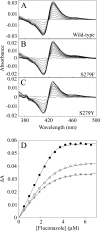
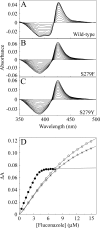
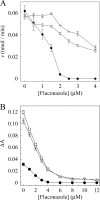
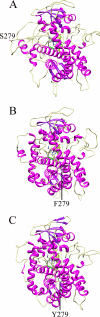
The effects of S279F and S279Y point mutations in Candida albicans CYP51 (CaCYP51) on protein activity and on substrate (lanosterol) and azole antifungal binding were investigated. Both S279F and S279Y mutants bound lanosterol with 2-fold increased affinities (K(s), 7.1 and 8.0 μM, respectively) compared to the wild-type CaCYP51 protein (K(s), 13.5 μM). The S279F and S279Y mutants and the wild-type CaCYP51 protein bound fluconazole, voriconazole, and itraconazole tightly, producing typical type II binding spectra. However, the S279F and S279Y mutants had 4- to 5-fold lower affinities for fluconazole, 3.5-fold lower affinities for voriconazole, and 3.5- to 4-fold lower affinities for itraconazole than the wild-type CaCYP51 protein. The S279F and S279Y mutants gave 2.3- and 2.8-fold higher 50% inhibitory concentrations (IC₅₀s) for fluconazole in a CYP51 reconstitution assay than the wild-type protein did. The increased fluconazole resistance conferred by the S279F and S279Y point mutations appeared to be mediated through a combination of a higher affinity for substrate and a lower affinity for fluconazole. In addition, lanosterol displaced fluconazole from the S279F and S279Y mutants but not from the wild-type protein. Molecular modeling of the wild-type protein indicated that the oxygen atom of S507 interacts with the second triazole ring of fluconazole, assisting in orientating fluconazole so that a more favorable binding conformation to heme is achieved. In contrast, in the two S279 mutant proteins, this S507-fluconazole interaction is absent, providing an explanation for the higher K(d) values observed.
Figures
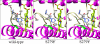
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Arase M, Waterman MR, Kagawa N. 2006. Purification and characterization of bovine steroid 21-hydroxylase (P450c21) efficiently expressed in Escherichia coli. Biochem. Biophys. Res. Commun. 344:400–405 - PubMed
-
- Bancel F, Bec N, Ebel C, Lange R. 1997. A central role for water in the control of the spin state of cytochrome P-450scc. Eur. J. Biochem. 250:276–285 - PubMed
-
- Bellamine A, Lepesheva GI, Waterman MR. 2004. Fluconazole binding and sterol demethylation in three CYP51 isoforms indicate differences in active site topology. J. Lipid Res. 45:2000–2007 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

