mRNA knockdown by single strand RNA is improved by chemical modifications
- PMID: 22253019
- PMCID: PMC3351186
- DOI: 10.1093/nar/gkr1301
mRNA knockdown by single strand RNA is improved by chemical modifications
Abstract
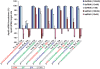
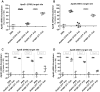
While RNAi has traditionally relied on RNA duplexes, early evaluation of siRNAs demonstrated activity of the guide strand in the absence of the passenger strand. However, these single strands lacked the activity of duplex RNAs. Here, we report the systematic use of chemical modifications to optimize single-strand RNA (ssRNA)-mediated mRNA knockdown. We identify that 2'F ribose modifications coupled with 5'-end phosphorylation vastly improves ssRNA activity both in vitro and in vivo. The impact of specific chemical modifications on ssRNA activity implies an Ago-mediated mechanism but the hallmark mRNA cleavage sites were not observed which suggests ssRNA may operate through a mechanism beyond conventional Ago2 slicer activity. While currently less potent than duplex siRNAs, with additional chemical optimization and alternative routes of delivery, chemically modified ssRNAs could represent a powerful RNAi platform.
Figures

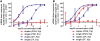


Similar articles
-
Argonaute-dependent small RNAs derived from single-stranded, non-structured precursors.Front Genet. 2014 Jun 10;5:172. doi: 10.3389/fgene.2014.00172. eCollection 2014. Front Genet. 2014. PMID: 24959173 Free PMC article. Review.
-
The 5' binding MID domain of human Argonaute2 tolerates chemically modified nucleotide analogues.Nucleic Acid Ther. 2013 Feb;23(1):81-7. doi: 10.1089/nat.2012.0393. Epub 2013 Jan 5. Nucleic Acid Ther. 2013. PMID: 23289589
-
Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5'-terminal phosphorylation both in vitro and in vivo.Nucleic Acids Res. 2010 Jan;38(2):660-71. doi: 10.1093/nar/gkp913. Epub 2009 Nov 16. Nucleic Acids Res. 2010. PMID: 19917641 Free PMC article.
-
Amide-Modified RNA: Using Protein Backbone to Modulate Function of Short Interfering RNAs.Acc Chem Res. 2020 Sep 15;53(9):1782-1790. doi: 10.1021/acs.accounts.0c00249. Epub 2020 Jul 13. Acc Chem Res. 2020. PMID: 32658452 Free PMC article.
-
Argonaute and Argonaute-Bound Small RNAs in Stem Cells.Int J Mol Sci. 2016 Feb 4;17(2):208. doi: 10.3390/ijms17020208. Int J Mol Sci. 2016. PMID: 26861290 Free PMC article. Review.
Cited by
-
Argonaute-dependent small RNAs derived from single-stranded, non-structured precursors.Front Genet. 2014 Jun 10;5:172. doi: 10.3389/fgene.2014.00172. eCollection 2014. Front Genet. 2014. PMID: 24959173 Free PMC article. Review.
-
Base modification strategies to modulate immune stimulation by an siRNA.Chembiochem. 2015 Jan 19;16(2):262-7. doi: 10.1002/cbic.201402551. Epub 2014 Dec 8. Chembiochem. 2015. PMID: 25487859 Free PMC article.
-
Optimization in Chemical Modification of Single-Stranded siRNA Encapsulated by Neutral Cytidinyl/Cationic Lipids.Front Chem. 2022 Mar 7;10:843181. doi: 10.3389/fchem.2022.843181. eCollection 2022. Front Chem. 2022. PMID: 35345539 Free PMC article.
-
ss-siRNAs allele selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy.Nucleic Acids Res. 2013 Nov;41(20):9570-83. doi: 10.1093/nar/gkt693. Epub 2013 Aug 9. Nucleic Acids Res. 2013. PMID: 23935115 Free PMC article.
-
Advances in modification and delivery of nucleic acid drugs.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2023 Aug 25;52(4):417-428. doi: 10.3724/zdxbyxb-2023-0130. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37643976 Free PMC article. Review. Chinese, English.
References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. - PubMed
-
- Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

