A gene regulatory network for root epidermis cell differentiation in Arabidopsis
- PMID: 22253603
- PMCID: PMC3257299
- DOI: 10.1371/journal.pgen.1002446
A gene regulatory network for root epidermis cell differentiation in Arabidopsis
Abstract
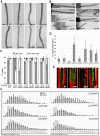
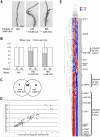
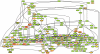
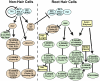
The root epidermis of Arabidopsis provides an exceptional model for studying the molecular basis of cell fate and differentiation. To obtain a systems-level view of root epidermal cell differentiation, we used a genome-wide transcriptome approach to define and organize a large set of genes into a transcriptional regulatory network. Using cell fate mutants that produce only one of the two epidermal cell types, together with fluorescence-activated cell-sorting to preferentially analyze the root epidermis transcriptome, we identified 1,582 genes differentially expressed in the root-hair or non-hair cell types, including a set of 208 "core" root epidermal genes. The organization of the core genes into a network was accomplished by using 17 distinct root epidermis mutants and 2 hormone treatments to perturb the system and assess the effects on each gene's transcript accumulation. In addition, temporal gene expression information from a developmental time series dataset and predicted gene associations derived from a Bayesian modeling approach were used to aid the positioning of genes within the network. Further, a detailed functional analysis of likely bHLH regulatory genes within the network, including MYC1, bHLH54, bHLH66, and bHLH82, showed that three distinct subfamilies of bHLH proteins participate in root epidermis development in a stage-specific manner. The integration of genetic, genomic, and computational analyses provides a new view of the composition, architecture, and logic of the root epidermal transcriptional network, and it demonstrates the utility of a comprehensive systems approach for dissecting a complex regulatory network.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Schellmann S, Hulskamp M, Uhrig J. Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem Soc Trans. 2007;35:146–148. - PubMed
-
- Grierson C, Schiefelbein J. Somerville C, Meyerowitz EM, editors. Root hairs. The Arabidopsis Book: American Society of Plant Biologists. 2002. http://www.aspb.org/publications/arabidopsis/
-
- Tominaga-Wada R, Ishida T, Wada T. New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int Rev Cell Mol Biol. 2011;286:67–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

