Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling
- PMID: 22253607
- PMCID: PMC3257278
- DOI: 10.1371/journal.pgen.1002452
Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling
Abstract
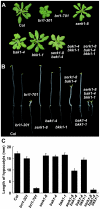
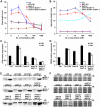
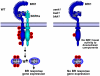
The Arabidopsis thaliana somatic embryogenesis receptor kinases (SERKs) consist of five members, SERK1 to SERK5, of the leucine-rich repeat receptor-like kinase subfamily II (LRR-RLK II). SERK3 was named BRI1-Associated Receptor Kinase 1 (BAK1) due to its direct interaction with the brassinosteroid (BR) receptor BRI1 in vivo, while SERK4 has also been designated as BAK1-Like 1 (BKK1) for its functionally redundant role with BAK1. Here we provide genetic and biochemical evidence to demonstrate that SERKs are absolutely required for early steps in BR signaling. Overexpression of four of the five SERKs-SERK1, SERK2, SERK3/BAK1, and SERK4/BKK1-suppressed the phenotypes of an intermediate BRI1 mutant, bri1-5. Overexpression of the kinase-dead versions of these four genes in the bri1-5 background, on the other hand, resulted in typical dominant negative phenotypes, resembling those of null BRI1 mutants. We isolated and generated single, double, triple, and quadruple mutants and analyzed their phenotypes in detail. While the quadruple mutant is embryo-lethal, the serk1 bak1 bkk1 triple null mutant exhibits an extreme de-etiolated phenotype similar to a null bri1 mutant. While overexpression of BRI1 can drastically increase hypocotyl growth of wild-type plants, overexpression of BRI1 does not alter hypocotyl growth of the serk1 bak1 bkk1 triple mutant. Biochemical analysis indicated that the phosphorylation level of BRI1 in serk1 bak1 bkk1 is incapable of sensing exogenously applied BR. As a result, the unphosphorylated level of BES1 has lost its sensitivity to the BR treatment in the triple mutant, indicating that the BR signaling pathway has been completely abolished in the triple mutant. These data clearly demonstrate that SERKs are essential to the early events of BR signaling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. - PubMed
-
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. - PubMed
-
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, et al. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713.
-
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

