Cytoplasmic polyadenylation element binding protein deficiency stimulates PTEN and Stat3 mRNA translation and induces hepatic insulin resistance
- PMID: 22253608
- PMCID: PMC3257279
- DOI: 10.1371/journal.pgen.1002457
Cytoplasmic polyadenylation element binding protein deficiency stimulates PTEN and Stat3 mRNA translation and induces hepatic insulin resistance
Abstract
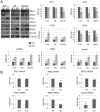
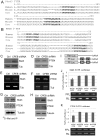
The cytoplasmic polyadenylation element binding protein CPEB1 (CPEB) regulates germ cell development, synaptic plasticity, and cellular senescence. A microarray analysis of mRNAs regulated by CPEB unexpectedly showed that several encoded proteins are involved in insulin signaling. An investigation of Cpeb1 knockout mice revealed that the expression of two particular negative regulators of insulin action, PTEN and Stat3, were aberrantly increased. Insulin signaling to Akt was attenuated in livers of CPEB-deficient mice, suggesting that they might be defective in regulating glucose homeostasis. Indeed, when the Cpeb1 knockout mice were fed a high-fat diet, their livers became insulin-resistant. Analysis of HepG2 cells, a human liver cell line, depleted of CPEB demonstrated that this protein directly regulates the translation of PTEN and Stat3 mRNAs. Our results show that CPEB regulated translation is a key process involved in insulin signaling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




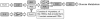
References
-
- White MF, Kahn CR. The insulin signaling system. J Biol Chem. 1994;269:1–4. - PubMed
-
- Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. - PubMed
-
- Kim JH, Kim JE, Liu HY, Cao W, Chen J. Regulation of interleukin-6-induced hepatic insulin resistance by mammalian target of rapamycin through the STAT3-SOCS3 pathway. J Biol Chem. 2008;283:708–715. - PubMed
-
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 2 T32 HD007312/HD/NICHD NIH HHS/United States
- P30 DK032520/DK/NIDDK NIH HHS/United States
- R01 AG030323/AG/NIA NIH HHS/United States
- 2 T32 HD007439/HD/NICHD NIH HHS/United States
- R01 HD037267/HD/NICHD NIH HHS/United States
- HD37267/HD/NICHD NIH HHS/United States
- R37 HD037267/HD/NICHD NIH HHS/United States
- AG30323/AG/NIA NIH HHS/United States
- DK32520/DK/NIDDK NIH HHS/United States
- T32 HD007312/HD/NICHD NIH HHS/United States
- U24 DK093000/DK/NIDDK NIH HHS/United States
- R01 GM046779/GM/NIGMS NIH HHS/United States
- R01 DK067493/DK/NIDDK NIH HHS/United States
- T32 HD007439/HD/NICHD NIH HHS/United States
- R01 DK080756/DK/NIDDK NIH HHS/United States
- DK80756/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

