Single base mismatches in the mRNA target site allow specific seed region-mediated off-target binding of siRNA targeting human coagulation factor 7
- PMID: 22258146
- PMCID: PMC3342946
- DOI: 10.4161/rna.9.1.18121
Single base mismatches in the mRNA target site allow specific seed region-mediated off-target binding of siRNA targeting human coagulation factor 7
Abstract
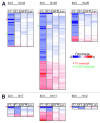
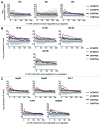
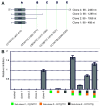
We have analyzed the off-target activity of two siRNAs (F7-1, F7-2) that knock-down human blood coagulation factor 7 mRNA. F7-1 modulates a significant number of non-target transcripts while F7-2 shows high selectivity for the target transcript under various experimental conditions. The 3'-UTRs of all F7-1 off-target genes show statistically significant enrichment of the reverse complement of the F7-1 siRNA seed region located in the guide strand. Seed region enrichment was confirmed in off-target transcripts modulated by siRNA targeting the glucocorticoid receptor. To investigate how these sites contribute to off-target recognition of F7-1, we employed CXCL5 transcript as model system because it contains five F7-1 seed sequence motifs with single base mismatches. We show by transient transfection of reporter gene constructs into HEK293 cells that three out of five sites located in the 3'-UTR region are required for F7-1 off-target activity. For further mechanistic dissection, the sequences of these sites were synthesized and inserted either individually or joined in dimeric or trimeric constructs. Only the fusion constructs were silenced by F7-1 while the individual sites had no off-target activity. Based on F7-1 as a model, a single mismatch between the siRNA seed region and mRNA target sites is tolerated for target recognition and the CXCL5 data suggest a requirement for binding to multiple target sites in off-target transcripts.
Figures
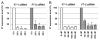


Similar articles
-
Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity.RNA. 2006 Jul;12(7):1179-87. doi: 10.1261/rna.25706. Epub 2006 May 8. RNA. 2006. PMID: 16682560 Free PMC article.
-
Experimental validation of the importance of seed complement frequency to siRNA specificity.RNA. 2008 May;14(5):853-61. doi: 10.1261/rna.704708. Epub 2008 Mar 26. RNA. 2008. PMID: 18367722 Free PMC article.
-
Modified siRNA structure with a single nucleotide bulge overcomes conventional siRNA-mediated off-target silencing.Mol Ther. 2011 Sep;19(9):1676-87. doi: 10.1038/mt.2011.109. Epub 2011 Jun 14. Mol Ther. 2011. PMID: 21673662 Free PMC article.
-
siRNA has greatly elevated mismatch tolerance at 3'-UTR sites.PLoS One. 2012;7(11):e49309. doi: 10.1371/journal.pone.0049309. Epub 2012 Nov 8. PLoS One. 2012. PMID: 23145149 Free PMC article.
-
Is the Efficiency of RNA Silencing Evolutionarily Regulated?Int J Mol Sci. 2016 May 12;17(5):719. doi: 10.3390/ijms17050719. Int J Mol Sci. 2016. PMID: 27187367 Free PMC article. Review.
Cited by
-
Downregulation of MicroRNA eca-mir-128 in Seminal Exosomes and Enhanced Expression of CXCL16 in the Stallion Reproductive Tract Are Associated with Long-Term Persistence of Equine Arteritis Virus.J Virol. 2018 Apr 13;92(9):e00015-18. doi: 10.1128/JVI.00015-18. Print 2018 May 1. J Virol. 2018. PMID: 29444949 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
