Structural changes in Influenza virus at low pH characterized by cryo-electron tomography
- PMID: 22258245
- PMCID: PMC3302323
- DOI: 10.1128/JVI.06698-11
Structural changes in Influenza virus at low pH characterized by cryo-electron tomography
Abstract
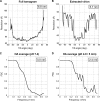
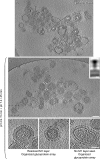
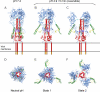
Influenza virus enters host cells by endocytosis. The low pH of endosomes triggers conformational changes in hemagglutinin (HA) that mediate fusion of the viral and endosomal membranes. We have used cryo-electron tomography to visualize influenza A virus at pH 4.9, a condition known to induce fusogenicity. After 30 min, when all virions are in the postfusion state, dramatic changes in morphology are apparent: elongated particles are no longer observed, larger particles representing fused virions appear, the HA spikes become conspicuously disorganized, a layer of M1 matrix protein is no longer resolved on most virions, and the ribonucleoprotein complexes (RNPs) coagulate on the interior surface of the virion. To probe for intermediate states, preparations were imaged after 5 min at pH 4.9. These virions could be classified according to their glycoprotein arrays (organized or disorganized) and whether or not they have a resolved M1 layer. Employing subtomogram averaging, we found, in addition to the neutral-pH state of HA, two intermediate conformations that appear to reflect an outwards movement of the fusion peptide and rearrangement of the HA1 subunits, respectively. These changes are reversible. The tomograms also document pH-induced changes affecting the M1 layer that appear to render the envelope more pliable and hence conducive to fusion. However, it appears desirable for productive infection that fusion should proceed before the RNPs become coagulated with matrix protein, as eventually happens at low pH.
Figures








References
-
- Barbey-Martin C, et al. 2002. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology 294:70–74 - PubMed
-
- Bizebard T, et al. 1995. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 376:92–94 - PubMed
-
- Booy FP, Ruigrok RW, van Bruggen EF. 1985. Electron microscopy of influenza virus. A comparison of negatively stained and ice-embedded particles. J. Mol. Biol. 184:667–676 - PubMed
-
- Bukrinskaya AG, Vorkunova NK, Kornilayeva GV, Narmanbetova RA, Vorkunova GK. 1982. Influenza virus uncoating in infected cells and effect of rimantadine. J. Gen. Virol. 60:49–59 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

