West nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response
- PMID: 22258263
- PMCID: PMC3302502
- DOI: 10.1128/JVI.06549-11
West nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response
Abstract
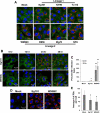
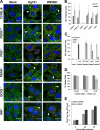
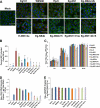
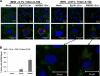
West Nile virus (WNV) recently became endemic in the United States and is a significant cause of human morbidity and mortality. Natural WNV strain infections do not induce stress granules (SGs), while W956IC (a lineage 2/1 chimeric WNV infectious clone) virus infections produce high levels of early viral RNA and efficiently induce SGs through protein kinase R (PKR) activation. Additional WNV chimeric viruses made by replacing one or more W956IC genes with the lineage 1 Eg101 equivalent in the W956IC backbone were analyzed. The Eg-NS4b+5, Eg-NS1+3+4a, and Eg-NS1+4b+5 chimeras produced low levels of viral RNA at early times of infection and inefficiently induced SGs, suggesting the possibility that interactions between viral nonstructural proteins and/or between viral nonstructural proteins and cell proteins are involved in suppressing early viral RNA synthesis and membrane remodeling during natural WNV strain infections. Detection of exposed viral double-stranded RNA (dsRNA) in W956IC-infected cells suggested that the enhanced early viral RNA synthesis surpassed the available virus-induced membrane protection and allowed viral dsRNA to activate PKR.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

