Disease gene identification strategies for exome sequencing
- PMID: 22258526
- PMCID: PMC3330229
- DOI: 10.1038/ejhg.2011.258
Disease gene identification strategies for exome sequencing
Abstract
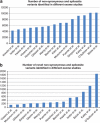
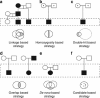
Next generation sequencing can be used to search for Mendelian disease genes in an unbiased manner by sequencing the entire protein-coding sequence, known as the exome, or even the entire human genome. Identifying the pathogenic mutation amongst thousands to millions of genomic variants is a major challenge, and novel variant prioritization strategies are required. The choice of these strategies depends on the availability of well-phenotyped patients and family members, the mode of inheritance, the severity of the disease and its population frequency. In this review, we discuss the current strategies for Mendelian disease gene identification by exome resequencing. We conclude that exome strategies are successful and identify new Mendelian disease genes in approximately 60% of the projects. Improvements in bioinformatics as well as in sequencing technology will likely increase the success rate even further. Exome sequencing is likely to become the most commonly used tool for Mendelian disease gene identification for the coming years.
Figures


References
-
- McKusick VA.Online Mendelian Inheritance in Man, OMIM http://www.ncbi.nlm.nih.gov/omim , http://www.ncbi.nlm.nih.gov/omim , 2011 - PubMed
-
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. - PubMed
-
- Mitchell KJ, Porteous DJ. Rethinking the genetic architecture of schizophrenia. Psychol Med. 2011;41:19–32. - PubMed
-
- Vissers LE, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

