Human cytomegalovirus transcriptome activity differs during replication in human fibroblast, epithelial and astrocyte cell lines
- PMID: 22258857
- PMCID: PMC3541802
- DOI: 10.1099/vir.0.038083-0
Human cytomegalovirus transcriptome activity differs during replication in human fibroblast, epithelial and astrocyte cell lines
Abstract
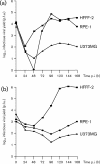
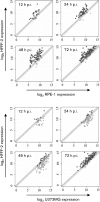
Broad cell tropism contributes to the pathogenesis of human cytomegalovirus (HCMV), but the extent to which cell type influences HCMV gene expression is unclear. A bespoke HCMV DNA microarray was used to monitor the transcriptome activity of the low passage Merlin strain of HCMV at 12, 24, 48 and 72 h post-infection, during a single round of replication in human fetal foreskin fibroblast cells (HFFF-2s), human retinal pigmented epithelial cells (RPE-1s) and human astrocytoma cells (U373MGs). In order to correlate transcriptome activity with concurrent biological responses, viral cytopathic effect, growth kinetics and genomic loads were examined in the three cell types. The temporal expression pattern of viral genes was broadly similar in HFFF-2s and RPE-1s, but dramatically different in U373MGs. Of the 165 known HCMV protein-coding genes, 41 and 48 were differentially regulated in RPE-1s and U373MGs, respectively, compared with HFFF-2s, and 22 of these were differentially regulated in both RPE-1s and U373MGs. In RPE-1s, all differentially regulated genes were downregulated, but, in U373MGs, some were down- and others upregulated. Differentially regulated genes were identified among the immediate-early, early, early late and true-late viral gene classes. Grouping of downregulated genes according to function at landmark stages of the replication cycle led to the identification of potential bottleneck stages (genome replication, virion assembly, and virion maturation and release) that may account for cell type-dependent viral growth kinetics. The possibility that cell type-specific differences in expressed cellular factors are responsible for modulation of viral gene expression is discussed.
Figures


Similar articles
-
Loss of the Human Cytomegalovirus US16 Protein Abrogates Virus Entry into Endothelial and Epithelial Cells by Reducing the Virion Content of the Pentamer.J Virol. 2017 May 12;91(11):e00205-17. doi: 10.1128/JVI.00205-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331097 Free PMC article.
-
Human cytomegalovirus temporally regulated gene expression in differentiated, immortalized retinal pigment epithelial cells.J Clin Virol. 2006 Apr;35(4):478-84. doi: 10.1016/j.jcv.2005.10.015. Epub 2006 Jan 4. J Clin Virol. 2006. PMID: 16388985
-
The US16 gene of human cytomegalovirus is required for efficient viral infection of endothelial and epithelial cells.J Virol. 2012 Jun;86(12):6875-88. doi: 10.1128/JVI.06310-11. Epub 2012 Apr 11. J Virol. 2012. PMID: 22496217 Free PMC article.
-
Coordination of late gene transcription of human cytomegalovirus with viral DNA synthesis: recombinant viruses as potential therapeutic vaccine candidates.Expert Opin Ther Targets. 2013 Feb;17(2):157-66. doi: 10.1517/14728222.2013.740460. Epub 2012 Dec 12. Expert Opin Ther Targets. 2013. PMID: 23231449 Review.
-
Human cytomegalovirus: taking the strain.Med Microbiol Immunol. 2015 Jun;204(3):273-84. doi: 10.1007/s00430-015-0411-4. Epub 2015 Apr 17. Med Microbiol Immunol. 2015. PMID: 25894764 Free PMC article. Review.
Cited by
-
Human Cytomegalovirus Productively Replicates In Vitro in Undifferentiated Oral Epithelial Cells.J Virol. 2018 Jul 31;92(16):e00903-18. doi: 10.1128/JVI.00903-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29848590 Free PMC article.
-
Subconfluent ARPE-19 Cells Display Mesenchymal Cell-State Characteristics and Behave like Fibroblasts, Rather Than Epithelial Cells, in Experimental HCMV Infection Studies.Viruses. 2023 Dec 28;16(1):49. doi: 10.3390/v16010049. Viruses. 2023. PMID: 38257749 Free PMC article.
-
New extracellular factors in glioblastoma multiforme development: neurotensin, growth differentiation factor-15, sphingosine-1-phosphate and cytomegalovirus infection.Oncotarget. 2018 Jan 9;9(6):7219-7270. doi: 10.18632/oncotarget.24102. eCollection 2018 Jan 23. Oncotarget. 2018. PMID: 29467963 Free PMC article. Review.
-
3D tissue-engineered lung models to study immune responses following viral infections of the small airways.Stem Cell Res Ther. 2022 Sep 7;13(1):464. doi: 10.1186/s13287-022-03134-1. Stem Cell Res Ther. 2022. PMID: 36071442 Free PMC article. Review.
-
Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation.PLoS Pathog. 2014 May 1;10(5):e1004058. doi: 10.1371/journal.ppat.1004058. eCollection 2014 May. PLoS Pathog. 2014. PMID: 24787765 Free PMC article.
References
-
- Aguilar J. S., Ghazal P., Wagner E. K. (2005). Design of a herpes simplex virus type 2 long oligonucleotide-based microarray: global analysis of HSV-2 transcript abundance during productive infection. Methods Mol Biol 292, 423–448 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

