Multilocus sequence typing and phylogenetic analysis of Propionibacterium acnes
- PMID: 22259216
- PMCID: PMC3318519
- DOI: 10.1128/JCM.r06129-11
Multilocus sequence typing and phylogenetic analysis of Propionibacterium acnes
Abstract
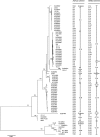
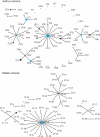
Propionibacterium acnes is a commensal of human skin but is also implicated in the pathogenesis of acne vulgaris, in biofilm-associated infections of medical devices and endophthalmitis, and in infections of bone and dental root canals. Recent studies associate P. acnes with prostate cancer. As the species includes evolutionary lineages with distinct association with health and disease, there is a need for a high-resolution typing scheme. Recently, two multilocus sequence typing (MLST) schemes were reported, one based on nine and one based on seven housekeeping genes. In the present study, the two schemes were compared with reference to a phylogenetic tree based on 78 P. acnes genomes and their gene contents. Further support for a basically clonal population structure of P. acnes and a scenario of the global spread of epidemic clones of P. acnes was obtained. Compared to the Belfast scheme, the Aarhus MLST scheme (http://pacnes.mlst.net/), which is based on nine genes, offers significantly enhanced resolution and phylogenetic inferences more concordant with analyses based on a comprehensive sampling of the entire genomes, their gene contents, and their putative pathogenic potential.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

