An in silico analysis of dynamic changes in microRNA expression profiles in stepwise development of nasopharyngeal carcinoma
- PMID: 22260379
- PMCID: PMC3293045
- DOI: 10.1186/1755-8794-5-3
An in silico analysis of dynamic changes in microRNA expression profiles in stepwise development of nasopharyngeal carcinoma
Abstract
Background: MicroRNAs (miRNAs) are small non-coding RNAs that participate in the spatiotemporal regulation of messenger RNA (mRNA) and protein synthesis. Recent studies have shown that some miRNAs are involved in the progression of nasopharyngeal carcinoma (NPC). However, the aberrant miRNAs implicated in different clinical stages of NPC remain unknown and their functions have not been systematically studied.
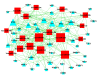
Methods: In this study, miRNA microarray assay was performed on biopsies from different clinical stages of NPC. TargetScan was used to predict the target genes of the miRNAs. The target gene list was narrowed down by searching the data from the UniGene database to identify the nasopharyngeal-specific genes. The data reduction strategy was used to overlay with nasopharyngeal-specifically expressed miRNA target genes and complementary DNA (cDNA) expression data. The selected target genes were analyzed in the Gene Ontology (GO) biological process and Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathway. The microRNA-Gene-Network was build based on the interactions of miRNAs and target genes. miRNA promoters were analyzed for the transcription factor (TF) binding sites. UCSC Genome database was used to construct the TF-miRNAs interaction networks.
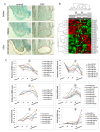
Results: Forty-eight miRNAs with significant change were obtained by Multi-Class Dif. The most enriched GO terms in the predicted target genes of miRNA were cell proliferation, cell migration and cell matrix adhesion. KEGG analysis showed that target genes were significantly involved in adherens junction, cell adhesion molecules, p53 signalling pathway et al. Comprehensive analysis of the coordinate expression of miRNAs and mRNAs reveals that miR-29a/c, miR-34b, miR-34c-3p, miR-34c-5p, miR-429, miR-203, miR-222, miR-1/206, miR-141, miR-18a/b, miR-544, miR-205 and miR-149 may play important roles on the development of NPC. We proposed an integrative strategy for identifying the miRNA-mRNA regulatory modules and TF-miRNA regulatory networks. TF including ETS2, MYB, Sp1, KLF6, NFE2, PCBP1 and TMEM54 exert regulatory functions on the miRNA expression.
Conclusions: This study provides perspective on the microRNA expression during the development of NPC. It revealed the global trends in miRNA interactome in NPC. It concluded that miRNAs might play important regulatory roles through the target genes and transcription factors in the stepwise development of NPC.
Figures

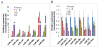
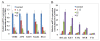
References
-
- Lin JC, Liao SK, Lee EH, Hung MS, Sayion Y, Chen HC, Kang CC, Huang LS, Cherng JM. Molecular events associated with epithelial to mesenchymal transition of nasopharyngeal carcinoma cells in the absence of Epstein-Barr virus genome. J Biomed Sci. 2009;16:105. doi: 10.1186/1423-0127-16-105. - DOI - PMC - PubMed
-
- Yoshizaki T, Ito M, Murono S, Wakisaka N, Kondo S, Endo K. Current understanding and management of nasopharyngeal carcinoma. Auris Nasus Larynx. 2011. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

