Post-natal cardiomyocytes can generate iPS cells with an enhanced capacity toward cardiomyogenic re-differentation
- PMID: 22261617
- PMCID: PMC3374080
- DOI: 10.1038/cdd.2011.205
Post-natal cardiomyocytes can generate iPS cells with an enhanced capacity toward cardiomyogenic re-differentation
Abstract
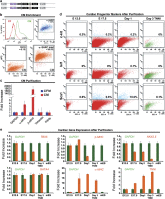
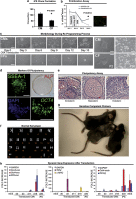
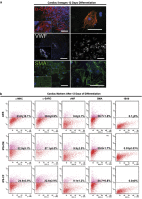
Adult mammalian cells can be reprogrammed to a pluripotent state by forcing the expression of a few embryonic transcription factors. The resulting induced pluripotent stem (iPS) cells can differentiate into cells of all three germ layers. It is well known that post-natal cardiomyocytes (CMs) lack the capacity to proliferate. Here, we report that neonatal CMs can be reprogrammed to generate iPS cells that express embryonic-specific markers and feature gene-expression profiles similar to those of mouse embryonic stem (mES) cell and cardiac fibroblast (CF)-derived iPS cell populations. CM-derived iPS cells are able to generate chimeric mice and, moreover, re-differentiate toward CMs more efficiently then either CF-derived iPS cells or mES cells. The increased differentiation capacity is possibly related to CM-derived iPS cells retaining an epigenetic memory of the phenotype of their founder cell. CM-derived iPS cells may thus lead to new information on differentiation processes underlying cardiac differentiation and proliferation.
Figures




Similar articles
-
Induced pluripotent stem cells expressing elevated levels of sox-2, oct-4, and klf-4 are severely reduced in their differentiation from mesodermal to hematopoietic progenitor cells.Stem Cells Dev. 2011 Jul;20(7):1131-42. doi: 10.1089/scd.2010.0391. Epub 2011 Feb 24. Stem Cells Dev. 2011. PMID: 21348597
-
Differentiation of reprogrammed mouse cardiac fibroblasts into functional cardiomyocytes.Cell Biochem Biophys. 2013 Jun;66(2):309-18. doi: 10.1007/s12013-012-9487-2. Cell Biochem Biophys. 2013. PMID: 23212180
-
Generation of induced pluripotent stem cells from neonatal mouse cochlear cells.Differentiation. 2014 Mar-Apr;87(3-4):127-33. doi: 10.1016/j.diff.2014.02.004. Epub 2014 Feb 28. Differentiation. 2014. PMID: 24582575
-
[Induction and characterization of induced pluripotent stem (iPS) cells: a review].Sheng Wu Gong Cheng Xue Bao. 2010 Apr;26(4):421-30. Sheng Wu Gong Cheng Xue Bao. 2010. PMID: 20575428 Review. Chinese.
-
iPS cells: a source of cardiac regeneration.J Mol Cell Cardiol. 2011 Feb;50(2):327-32. doi: 10.1016/j.yjmcc.2010.10.026. Epub 2010 Oct 30. J Mol Cell Cardiol. 2011. PMID: 21040726 Review.
Cited by
-
Cardiac Fibroblast-Induced Pluripotent Stem Cell-Derived Exosomes as a Potential Therapeutic Mean for Heart Failure.Int J Mol Sci. 2020 Sep 29;21(19):7215. doi: 10.3390/ijms21197215. Int J Mol Sci. 2020. PMID: 33003641 Free PMC article.
-
Reprogramming-derived gene cocktail increases cardiomyocyte proliferation for heart regeneration.EMBO Mol Med. 2017 Feb;9(2):251-264. doi: 10.15252/emmm.201606558. EMBO Mol Med. 2017. PMID: 28011860 Free PMC article.
-
A multi-cellular 3D bioprinting approach for vascularized heart tissue engineering based on HUVECs and iPSC-derived cardiomyocytes.Sci Rep. 2018 Sep 10;8(1):13532. doi: 10.1038/s41598-018-31848-x. Sci Rep. 2018. PMID: 30201959 Free PMC article.
-
Genetically matched human iPS cells reveal that propensity for cartilage and bone differentiation differs with clones, not cell type of origin.PLoS One. 2013;8(1):e53771. doi: 10.1371/journal.pone.0053771. Epub 2013 Jan 31. PLoS One. 2013. PMID: 23382851 Free PMC article.
-
A Concise Review on Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Personalized Regenerative Medicine.Stem Cell Rev Rep. 2021 Jun;17(3):748-776. doi: 10.1007/s12015-020-10061-2. Epub 2020 Oct 23. Stem Cell Rev Rep. 2021. PMID: 33098306 Review.
References
-
- Murry CE, Lee RT. Development biology. Turnover after the fallout. Science. 2009;324:47–48. - PubMed
-
- Krause K, Schneider C, Kuck KH, Jaquet K. Stem cell therapy in cardiovascular disorders. Cardiovasc Ther. 2010;28:e101–e110. - PubMed
-
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. - PubMed
-
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

