Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture
- PMID: 22261722
- PMCID: PMC3433907
- DOI: 10.1074/mcp.M111.014233
Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture
Abstract
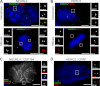
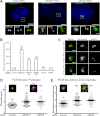
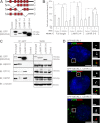
Centrosomes are composed of a centriole pair surrounded by an intricate proteinaceous matrix referred to as pericentriolar material. Although the mechanisms underpinning the control of centriole duplication are now well understood, we know relatively little about the control of centrosome size and shape. Here we used interaction proteomics to identify the E3 ligase HERC2 and the neuralized homologue NEURL4 as novel interaction partners of the centrosomal protein CP110. Using high resolution imaging, we find that HERC2 and NEURL4 localize to the centrosome and that interfering with their function alters centrosome morphology through the appearance of aberrant filamentous structures that stain for a subset of pericentriolar material proteins including pericentrin and CEP135. Using an RNA interference-resistant transgene approach in combination with structure-function analyses, we show that the association between CP110 and HERC2 depends on nonoverlapping regions of NEURL4. Whereas CP110 binding to NEURL4 is dispensable for the regulation of pericentriolar material architecture, its association with HERC2 is required to maintain normal centrosome integrity. NEURL4 is a substrate of HERC2, and together these results indicate that the NEURL4-HERC2 complex participates in the ubiquitin-dependent regulation of centrosome architecture.
Figures


Similar articles
-
Neurl4, a novel daughter centriole protein, prevents formation of ectopic microtubule organizing centres.EMBO Rep. 2012 Jun 1;13(6):547-53. doi: 10.1038/embor.2012.40. EMBO Rep. 2012. PMID: 22441691 Free PMC article.
-
NudC-like protein 2 restrains centriole amplification by stabilizing HERC2.Cell Death Dis. 2019 Aug 19;10(9):628. doi: 10.1038/s41419-019-1843-3. Cell Death Dis. 2019. PMID: 31427565 Free PMC article.
-
Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes.Mol Cell Biol. 2012 Aug;32(15):3095-106. doi: 10.1128/MCB.00201-12. Epub 2012 May 29. Mol Cell Biol. 2012. PMID: 22645313 Free PMC article.
-
Centrosomes in the DNA damage response--the hub outside the centre.Chromosome Res. 2016 Jan;24(1):35-51. doi: 10.1007/s10577-015-9503-7. Chromosome Res. 2016. PMID: 26614090 Review.
-
A centrosomal scaffold shows some self-control.J Biol Chem. 2017 Dec 15;292(50):20410-20411. doi: 10.1074/jbc.H117.806018. J Biol Chem. 2017. PMID: 29247130 Free PMC article. Review.
Cited by
-
Insights into the ubiquitin-proteasome system of human embryonic stem cells.Sci Rep. 2018 Mar 6;8(1):4092. doi: 10.1038/s41598-018-22384-9. Sci Rep. 2018. PMID: 29511261 Free PMC article.
-
Neuralized-like protein 4 (NEURL4) mediates ADP-ribosylation of mitochondrial proteins.J Cell Biol. 2022 Mar 7;221(3):e202101021. doi: 10.1083/jcb.202101021. Epub 2022 Feb 14. J Cell Biol. 2022. PMID: 35157000 Free PMC article.
-
CDK2 Inhibition Causes Anaphase Catastrophe in Lung Cancer through the Centrosomal Protein CP110.Cancer Res. 2015 May 15;75(10):2029-38. doi: 10.1158/0008-5472.CAN-14-1494. Epub 2015 Mar 25. Cancer Res. 2015. PMID: 25808870 Free PMC article.
-
Functional and pathological relevance of HERC family proteins: a decade later.Cell Mol Life Sci. 2016 May;73(10):1955-68. doi: 10.1007/s00018-016-2139-8. Epub 2016 Jan 22. Cell Mol Life Sci. 2016. PMID: 26801221 Free PMC article. Review.
-
Coordination of Embryogenesis by the Centrosome in Drosophila melanogaster.Results Probl Cell Differ. 2019;67:277-321. doi: 10.1007/978-3-030-23173-6_12. Results Probl Cell Differ. 2019. PMID: 31435800 Free PMC article. Review.
References
-
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 - PubMed
-
- Nigg E. A., Raff J. W. (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 - PubMed
-
- Palazzo R. E., Vogel J. M., Schnackenberg B. J., Hull D. R., Wu X. (2000) Centrosome maturation. Curr. Top. Dev. Biol. 49, 449–470 - PubMed
-
- Conduit P. T., Brunk K., Dobbelaere J., Dix C. I., Lucas E. P., Raff J. W. (2010) Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 20, 2178–2186 - PubMed
-
- Decker M., Jaensch S., Pozniakovsky A., Zinke A., O'Connell K. F., Zachariae W., Myers E., Hyman A. A. (2011) Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 21, 1259–1267 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

