Physical characterization of the "immunosignaturing effect"
- PMID: 22261726
- PMCID: PMC3367934
- DOI: 10.1074/mcp.M111.011593
Physical characterization of the "immunosignaturing effect"
Abstract
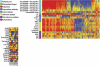
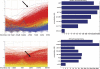
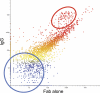
Identifying new, effective biomarkers for diseases is proving to be a challenging problem. We have proposed that antibodies may offer a solution to this problem. The physical features and abundance of antibodies make them ideal biomarkers. Additionally, antibodies are often elicited early in the ontogeny of different chronic and infectious diseases. We previously reported that antibodies from patients with infectious disease and separately those with Alzheimer's disease display a characteristic and reproducible "immunosignature" on a microarray of 10,000 random sequence peptides. Here we investigate the physical and chemical parameters underlying how immunosignaturing works. We first show that a variety of monoclonal and polyclonal antibodies raised against different classes of antigens produce distinct profiles on this microarray and the relative affinities are determined. A proposal for how antibodies bind the random sequences is tested. Sera from vaccinated mice and people suffering from a fugal infection are individually assayed to determine the complexity of signals that can be distinguished. Based on these results, we propose that this simple, general and inexpensive system could be optimized to generate a new class of antibody biomarkers for a wide variety of diseases.
Figures







References
-
- Kurian S. M., Heilman R., Mondala T. S., Nakorchevsky A., Hewel J. A., Campbell D., Robison E. H., Wang L., Lin W., Gaber L., Solez K., Shidban H., Mendez R., Schaffer R. L., Fisher J. S., Flechner S. M., Head S. R., Horvath S., Yates J. R., Marsh C. L., Salomon D. R. (2009) Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS ONE 4, e6212. - PMC - PubMed
-
- Amur S., Frueh F. W., Lesko L. J., Huang S. M. (2008) Integration and use of biomarkers in drug development, regulation and clinical practice: a US regulatory perspective. Biomarkers Med. 2, 305–311 - PubMed
-
- Food and Drug Administration (2010) Table of valid genomic biomarkers in the context of approved drug labels. http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/...
-
- Sulzer B., van Hemmen J. L., Neumann A.U., Behn U. (1993) Memory in idiotypic networks due to competition between proliferation and differentiation. Bull. Math. Biol. 55, 1133–1182 - PubMed
-
- Cenci S., Sitia R. (2007) Managing and exploiting stress in the antibody factory. FEBS Lett. 581, 3652–3657 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

