Solution structure analysis of cytoplasmic domain of podocyte protein Neph1 using small/wide angle x-ray scattering (SWAXS)
- PMID: 22262837
- PMCID: PMC3308819
- DOI: 10.1074/jbc.M111.284927
Solution structure analysis of cytoplasmic domain of podocyte protein Neph1 using small/wide angle x-ray scattering (SWAXS)
Abstract
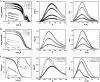
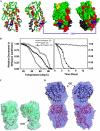
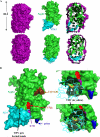
Neph1 is present in podocytes, where it plays a critical role in maintaining the filtration function of the glomerulus, in part through signaling events mediated by its cytoplasmic domain that are involved in actin cytoskeleton organization. To understand the function of this protein, a detailed knowledge of the structure of the Neph1 cytoplasmic domain (Neph1-CD) is required. In this study, the solution structure of this domain was determined by small/wide angle x-ray scattering (SWAXS). Analysis of Neph1-CD by SWAXS suggested that this protein adopts a global shape with a radius of gyration and a maximum linear dimension of 21.3 and 70 Å, respectively. These parameters and the data from circular dichroism experiments were used to construct a structural model of this protein. The His-ZO-1-PDZ1 (first PDZ domain of zonula occludens) domain that binds Neph1-CD was also analyzed by SWAXS, to confirm that it adopts a global structure similar to its crystal structure. We used the SWAXS intensity profile, the structural model of Neph1-CD, and the crystal structure of ZO-1-PDZ1 to construct a structural model of the Neph1-CD·ZO-1-PDZ1 complex. Mapping of the intermolecular interactions suggested that in addition to the C-terminal residues Thr-His-Val, residues Lys-761 and Tyr-762 in Neph1 are also critical for stabilizing the complex. Estimated intensity values from the SWAXS data and in vivo and in vitro pull-down experiments demonstrated loss of binding to ZO-1 when these residues were individually mutated to alanines. Our findings present a structural model that provides novel insights into the molecular structure and function of Neph1-CD.
Figures



References
-
- Patrakka J., Tryggvason K. (2010) Molecular make-up of the glomerular filtration barrier. Biochem. Biophys. Res. Commun. 396, 164–169 - PubMed
-
- Tryggvason K., Pikkarainen T., Patrakka J. (2006) Nck links nephrin to actin in kidney podocytes. Cell 125, 221–224 - PubMed
-
- Moreno J. A., Sanchez-Niño M. D., Sanz A. B., Lassila M., Holthofer H., Blanco-Colio L. M., Egido J., Ruiz-Ortega M., Ortiz A. (2008) A slit in podocyte death. Curr. Med. Chem. 15, 1645–1654 - PubMed
-
- Johnstone D. B., Holzman L. B. (2006) Clinical impact of research on the podocyte slit diaphragm. Nat. Clin. Pract. Nephrol. 2, 271–282 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

