Combinatorial expression of Brn3 transcription factors in somatosensory neurons: genetic and morphologic analysis
- PMID: 22262898
- PMCID: PMC3428801
- DOI: 10.1523/JNEUROSCI.4755-11.2012
Combinatorial expression of Brn3 transcription factors in somatosensory neurons: genetic and morphologic analysis
Abstract
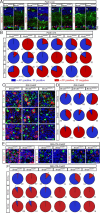
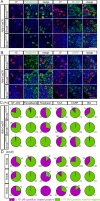
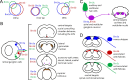
The three members of the Brn3 family of POU-domain transcription factors (Brn3a/Pou4f1, Brn3b/Pou4f2, and Brn3c/Pou4f3) are expressed in overlapping subsets of visual, auditory/vestibular, and somatosensory neurons. Using unmarked Brn3-null alleles and Brn3 conditional alleles in which gene loss is coupled to expression of an alkaline phosphatase reporter, together with sparse Cre-mediated recombination, we describe the following: (1) the overlapping patterns of Brn3 gene expression in somatosensory neurons; (2) the manner in which these patterns correlate with molecular markers, peripheral afferent arbor morphologies, and dorsal horn projections; and (3) the consequences for these neurons of deleting individual Brn3 genes in the mouse. We observe broad expression of Brn3a among DRG neurons, but subtype-restricted expression of Brn3b and Brn3c. We also observe a nearly complete loss of hair follicle-associated sensory endings among Brn3a(-/-) neurons. Together with earlier analyses of Brn3 gene expression patterns in the retina and inner ear, these experiments suggest a deep functional similarity among primary somatosensory neurons, spiral and vestibular ganglion neurons, and retinal ganglion cells. This work also demonstrates the utility of sparse genetically directed labeling for visualizing individual somatosensory afferent arbors and for defining cell-autonomous mutant phenotypes.
Figures





References
-
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480:331–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
