Understanding the language of Lys36 methylation at histone H3
- PMID: 22266761
- PMCID: PMC3969746
- DOI: 10.1038/nrm3274
Understanding the language of Lys36 methylation at histone H3
Abstract
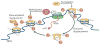
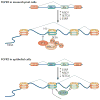
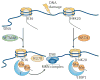
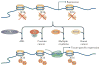
Histone side chains are post-translationally modified at multiple sites, including at Lys36 on histone H3 (H3K36). Several enzymes from yeast and humans, including the methyltransferases SET domain-containing 2 (Set2) and nuclear receptor SET domain-containing 1 (NSD1), respectively, alter the methylation status of H3K36, and significant progress has been made in understanding how they affect chromatin structure and function. Although H3K36 methylation is most commonly associated with the transcription of active euchromatin, it has also been implicated in diverse processes, including alternative splicing, dosage compensation and transcriptional repression, as well as DNA repair and recombination. Disrupted placement of methylated H3K36 within the chromatin landscape can lead to a range of human diseases, underscoring the importance of this modification.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Allfrey VG, Mirsky AE. Structural modifications of histones and their possible role in the regulation of RNA Synthesis. Science. 1964;144:559. - PubMed
-
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature Rev Genet. 2006;7:715–727. - PubMed
-
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2011;1815:75–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

