The miR-17-92 cluster regulates FOG-2 expression and inhibits proliferation of mouse embryonic cardiomyocytes
- PMID: 22267003
- PMCID: PMC3854259
- DOI: 10.1590/s0100-879x2012007500007
The miR-17-92 cluster regulates FOG-2 expression and inhibits proliferation of mouse embryonic cardiomyocytes
Abstract
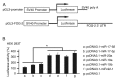
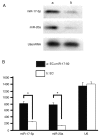
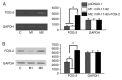
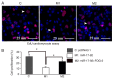
MicroRNAs (miRNAs) have gradually been recognized as regulators of embryonic development; however, relatively few miRNAs have been identified that regulate cardiac development. A series of recent papers have established an essential role for the miRNA-17-92 (miR-17-92) cluster of miRNAs in the development of the heart. Previous research has shown that the Friend of Gata-2 (FOG-2) is critical for cardiac development. To investigate the possibility that the miR-17-92 cluster regulates FOG-2 expression and inhibits proliferation in mouse embryonic cardiomyocytes we initially used bioinformatics to analyze 3' untranslated regions (3'UTR) of FOG-2 to predict the potential of miR-17-92 to target it. We used luciferase assays to demonstrate that miR-17-5p and miR-20a of miR-17-92 interact with the predicted target sites in the 3'UTR of FOG-2. Furthermore, RT-PCR and Western blot were used to demonstrate the post-transcriptional regulation of FOG-2 by miR-17-92 in embryonic cardiomyocytes from E12.5-day pregnant C57BL/6J mice. Finally, EdU cell assays together with the FOG-2 rescue strategy were employed to evaluate the effect of proliferation on embryonic cardiomyocytes. We first found that the miR-17-5p and miR-20a of miR-17-92 directly target the 3'UTR of FOG-2 and post-transcriptionally repress the expression of FOG-2. Moreover, our findings demonstrated that over-expression of miR-17-92 may inhibit cell proliferation via post-transcriptional repression of FOG-2 in embryonic cardiomyocytes. These results indicate that the miR-17-92 cluster regulates the expression of FOG-2 protein and suggest that the miR-17-92 cluster might play an important role in heart development.
Figures

References
-
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

