Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium
- PMID: 22267480
- PMCID: PMC3306477
- DOI: 10.1161/ATVBAHA.111.244053
Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium
Abstract
Objective: Endothelial transcription factors Krüppel-like factor 4 (KLF4) and KLF2 are implicated in protection against atherogenesis. Steady-state microRNA (miR) regulation of KLFs in vivo is accessible by screening region-specific endothelial miRs and their targets.
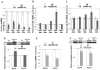
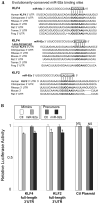
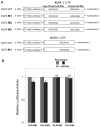
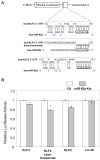
Methods and results: A subset of differentially expressed endothelial miRs was identified in atherosusceptible versus protected regions of normal swine aorta. In silico analyses predicted highly conserved binding sites in the 3'-untranslated region (3'UTR) of KLF4 for 5 miRs of the subset (miR-26a, -26b, -29a, -92a, and -103) and a single binding site for a miR-92a complex in the 3'UTR of KLF2. Of these, only miR-92a knockdown and knock-in resulted in responses of KLF4 and KLF2 expression in human arterial endothelial cells. Dual luciferase reporter assays demonstrated functional interactions of miR-92a with full-length 3'UTR sequences of both KLFs and with the specific binding elements therein. Two evolutionarily conserved miR-92a sites in KLF4 3'UTR and 1 site in KLF2 3'UTR were functionally validated. Knockdown of miR-92a in vitro resulted in partial rescue from cytokine-induced proinflammatory marker expression (monocyte chemotactic protein 1, vascular cell adhesion molecule-1, E-selectin, and endothelial nitric oxide synthase) that was attributable to enhanced KLF4 expression. Leukocyte-human arterial endothelial cell adhesion experiments supported this conclusion. In swine aortic arch endothelium, a site of atherosusceptibility where miR-92a expression was elevated, both KLFs were expressed at low levels relative to protected thoracic aorta.
Conclusions: miR-92a coregulates KLF4 and KLF2 expression in arterial endothelium and contributes to phenotype heterogeneity associated with regional atherosusceptibility and protection in vivo.
Conflict of interest statement
(c) The authors have no conflicts to disclose.
Figures


Comment in
-
MiRrored regulation of KLF2 and KLF4.Arterioscler Thromb Vasc Biol. 2012 Apr;32(4):839-40. doi: 10.1161/ATVBAHA.112.245563. Arterioscler Thromb Vasc Biol. 2012. PMID: 22423032 Free PMC article. No abstract available.
References
-
- Passerini AG, Polacek DC, Shi C, Francesco NM, Manduchi E, Grant GR, Pritchard WF, Powell S, Chang GY, Stoeckert CJ, Jr, Davies PF. Coexisting proinflammatory and antioxidative endothelial transcription profiles in a disturbed flow region of the adult porcine aorta. Proc Natl Acad Sci U S A. 2004;101:2482–2487. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

