Biochemical inhibition of the acetyltransferases ATase1 and ATase2 reduces β-secretase (BACE1) levels and Aβ generation
- PMID: 22267734
- PMCID: PMC3318698
- DOI: 10.1074/jbc.M111.310136
Biochemical inhibition of the acetyltransferases ATase1 and ATase2 reduces β-secretase (BACE1) levels and Aβ generation
Abstract
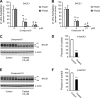
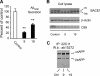
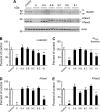
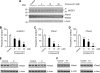
The cellular levels of β-site APP cleaving enzyme 1 (BACE1), the rate-limiting enzyme for the generation of the Alzheimer disease (AD) amyloid β-peptide (Aβ), are tightly regulated by two ER-based acetyl-CoA:lysine acetyltransferases, ATase1 and ATase2. Here we report that both acetyltransferases are expressed in neurons and glial cells, and are up-regulated in the brain of AD patients. We also report the identification of first and second generation compounds that inhibit ATase1/ATase2 and down-regulate the expression levels as well as activity of BACE1. The mechanism of action involves competitive and non-competitive inhibition as well as generation of unstable intermediates of the ATases that undergo degradation.
Figures





References
-
- Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., Klein W. L. (1998) Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 - PMC - PubMed
-
- Klein W. L., Krafft G. A., Finch C. E. (2001) Targeting small Aβ oligomers: the solution to an Alzheimer disease conundrum? Trends Neurosci. 24, 219–224 - PubMed
-
- Haass C., Steiner H. (2001) Protofibrils, the unifying toxic molecule of neurodegenerative disorders? Nat. Neurosci. 4, 859–860 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

