Assisted reproduction treatment and epigenetic inheritance
- PMID: 22267841
- PMCID: PMC3282574
- DOI: 10.1093/humupd/dmr047
Assisted reproduction treatment and epigenetic inheritance
Abstract
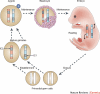
Background: The subject of epigenetic risk of assisted reproduction treatment (ART), initiated by reports on an increase of children with the Beckwith-Wiedemann imprinting disorder, is very topical. Hence, there is a growing literature, including mouse studies.
Methods: In order to gain information on transgenerational epigenetic inheritance and epigenetic effects induced by ART, literature databases were searched for papers on this topic using relevant keywords.
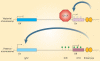
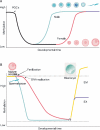
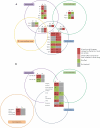
Results: At the level of genomic imprinting involving CpG methylation, ART-induced epigenetic defects are convincingly observed in mice, especially for placenta, and seem more frequent than in humans. Data generally provide a warning as to the use of ovulation induction and in vitro culture. In human sperm from compromised spermatogenesis, sequence-specific DNA hypomethylation is observed repeatedly. Transmittance of sperm and oocyte DNA methylation defects is possible but, as deduced from the limited data available, largely prevented by selection of gametes for ART and/or non-viability of the resulting embryos. Some evidence indicates that subfertility itself is a risk factor for imprinting diseases. As in mouse, physiological effects from ART are observed in humans. In the human, indications for a broader target for changes in CpG methylation than imprinted DNA sequences alone have been found. In the mouse, a broader range of CpG sequences has not yet been studied. Also, a multigeneration study of systematic ART on epigenetic parameters is lacking.
Conclusions: The field of epigenetic inheritance within the lifespan of an individual and between generations (via mitosis and meiosis, respectively) is growing, driven by the expansion of chromatin research. ART can induce epigenetic variation that might be transmitted to the next generation.
Figures


References
-
- Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23:2826–2834. doi:10.1093/humrep/den310. - DOI - PubMed
-
- Anderson LM, Riffle L, Wilson R, Travlos GS, Lubomirski MS, Alvord WG. Preconceptional fasting of fathers alters serum glucose in offspring of mice. Nutrition. 2006;22:327–331. doi:10.1016/j.nut.2005.09.006. - DOI - PubMed
-
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and mate fertility. Science. 2005;308:1466–1469. doi:10.1126/science.1108190. - DOI - PMC - PubMed
-
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi:10.1016/j.molcel.2008.09.003. - DOI - PMC - PubMed
-
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic Compartmentalization of the Fetal piRNA Pathway in Mice. Plos Genet. 2009;5:12. doi:10.1371/journal.pgen.1000764. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

