WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana
- PMID: 22268143
- PMCID: PMC3346227
- DOI: 10.1093/jxb/err450
WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana
Abstract
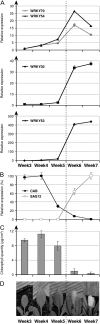
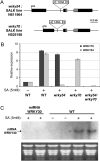
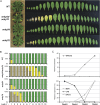
The plant-specific WRKY transcription factor (TF) family with 74 members in Arabidopsis thaliana appears to be involved in the regulation of various physiological processes including plant defence and senescence. WRKY53 and WRKY70 were previously implicated as positive and negative regulators of senescence, respectively. Here the putative function of other WRKY group III proteins in Arabidopsis leaf senescence has been explored and the results suggest the involvement of two additional WRKY TFs, WRKY 54 and WRKY30, in this process. The structurally related WRKY54 and WRKY70 exhibit a similar expression pattern during leaf development and appear to have co-operative and partly redundant functions in senescence, as revealed by single and double mutant studies. These two negative senescence regulators and the positive regulator WRKY53 were shown by yeast two-hydrid analysis to interact independently with WRKY30. WRKY30 was expressed during developmental leaf senescence and consequently it is hypothesized that the corresponding protein could participate in a senescence regulatory network with the other WRKYs. Expression in wild-type and salicylic acid-deficient mutants suggests a common but not exclusive role for SA in induction of WRKY30, 53, 54, and 70 during senescence. WRKY30 and WRKY53 but not WRKY54 and WRKY70 are also responsive to additional signals such as reactive oxygen species. The results suggest that WRKY53, WRKY54, and WRKY70 may participate in a regulatory network that integrates internal and environmental cues to modulate the onset and the progression of leaf senescence, possibly through an interaction with WRKY30.
Figures



References
-
- AbuQamar S, Chen X, Dhawan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T. Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. The Plant Journal. 2006;48:28–44. - PubMed
-
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biology (Stuttgart) 2008;1(10 Suppl):63–75. - PubMed
-
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnology Journal. 2003;1:3–22. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

