Characterization of the C. elegans erlin homologue
- PMID: 22269071
- PMCID: PMC3292932
- DOI: 10.1186/1471-2121-13-2
Characterization of the C. elegans erlin homologue
Abstract
Background: Erlins are highly conserved proteins associated with lipid rafts within the endoplasmic reticulum (ER). Biochemical studies in mammalian cell lines have shown that erlins are required for ER associated protein degradation (ERAD) of activated inositol-1,4,5-trisphosphate receptors (IP3Rs), implying that erlin proteins might negatively regulate IP3R signalling. In humans, loss of erlin function appears to cause progressive intellectual disability, motor dysfunction and joint contractures. However, it is unknown if defects in IP3R ERAD are the underlying cause of this disease phenotype, whether ERAD of activated IP3Rs is the only function of erlin proteins, and what role ERAD plays in regulating IP3R-dependent processes in the context of an intact animal or embryo. In this study, we characterize the erlin homologue of the nematode Caenorhabditis elegans and examine erlin function in vivo. We specifically set out to test whether C. elegans erlin modulates IP3R-dependent processes, such as egg laying, embryonic development and defecation rates. We also explore the possibility that erlin might play a more general role in the ERAD pathway of C. elegans.
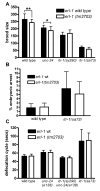
Results: We first show that the C. elegans erlin homologue, ERL-1, is highly similar to mammalian erlins with respect to amino acid sequence, domain structure, biochemical properties and subcellular location. ERL-1 is present throughout the C. elegans embryo; in adult worms, ERL-1 appears restricted to the germline. The expression pattern of ERL-1 thus only partially overlaps with that of ITR-1, eliminating the possibility of ERL-1 being a ubiquitous and necessary regulator of ITR-1. We show that loss of ERL-1 does not affect overall phenotype, or alter brood size, embryonic development or defecation cycle length in either wild type or sensitized itr-1 mutant animals. Moreover we show that ERL-1 deficient worms respond normally to ER stress conditions, suggesting that ERL-1 is not an essential component of the general ERAD pathway.
Conclusions: Although loss of erlin function apparently causes a strong phenotype in humans, no such effect is seen in C. elegans. C. elegans erlin does not appear to be a ubiquitous major modulator of IP3 receptor activity nor does erlin appear to play a major role in ERAD.
Figures






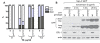
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

