La-motif-dependent mRNA association with Slf1 promotes copper detoxification in yeast
- PMID: 22271760
- PMCID: PMC3285933
- DOI: 10.1261/rna.028506.111
La-motif-dependent mRNA association with Slf1 promotes copper detoxification in yeast
Abstract
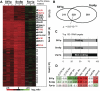
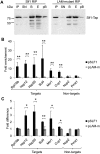
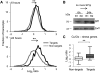
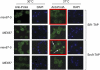
The La-motif (LAM) is an ancient and ubiquitous RNA-binding domain defining a superfamily of proteins, which comprises the genuine La proteins and La-related proteins (LARPs). In contrast to La, which binds and stabilizes pre-tRNAs and other RNA polymerase III transcripts, data on function and RNA targets of the LARPs have remained scarce. We have undertaken a global approach to elucidate the previously suggested role of the yeast LARP Slf1p in copper homeostasis. By applying RNA-binding protein immunopurification-microarray (RIP-Chip) analysis, we show that Slf1p and its paralog Sro9p copurify with overlapping sets of hundreds of functionally related mRNAs, including many transcripts coding for ribosomal proteins and histones. Interestingly, among these potential RNA targets were also mRNAs coding for proteins critical for protection of cells against elevated copper concentrations. Mutations introduced in the conserved aromatic patch of the LAM in Slf1p drastically impaired both association with its targets and Slf1-mediated protection of cells against toxic copper concentrations. Furthermore, we show that Slf1p stabilizes copper-related mRNA targets in a LAM-dependent manner. These results provide the first evidence for post-transcriptional regulation of factors/pathways implicated in copper homeostasis by a cytoplasmic RBP.
Figures


Similar articles
-
The yeast La related protein Slf1p is a key activator of translation during the oxidative stress response.PLoS Genet. 2015 Jan 8;11(1):e1004903. doi: 10.1371/journal.pgen.1004903. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569619 Free PMC article.
-
Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes.Mol Biol Cell. 1999 Nov;10(11):3849-62. doi: 10.1091/mbc.10.11.3849. Mol Biol Cell. 1999. PMID: 10564276 Free PMC article.
-
Conserved and divergent features of the structure and function of La and La-related proteins (LARPs).Biochim Biophys Acta. 2010 May-Jun;1799(5-6):365-78. doi: 10.1016/j.bbagrm.2010.01.011. Epub 2010 Feb 2. Biochim Biophys Acta. 2010. PMID: 20138158 Free PMC article. Review.
-
A comprehensive analysis of the La-motif protein superfamily.RNA. 2009 May;15(5):750-64. doi: 10.1261/rna.1478709. Epub 2009 Mar 19. RNA. 2009. PMID: 19299548 Free PMC article.
-
The role of LARP1 in translation and beyond.Wiley Interdiscip Rev RNA. 2015 Jul-Aug;6(4):399-417. doi: 10.1002/wrna.1282. Epub 2015 Apr 17. Wiley Interdiscip Rev RNA. 2015. PMID: 25892282 Review.
Cited by
-
Puf3p induces translational repression of genes linked to oxidative stress.Nucleic Acids Res. 2014 Jan;42(2):1026-41. doi: 10.1093/nar/gkt948. Epub 2013 Oct 25. Nucleic Acids Res. 2014. PMID: 24163252 Free PMC article.
-
The yeast La related protein Slf1p is a key activator of translation during the oxidative stress response.PLoS Genet. 2015 Jan 8;11(1):e1004903. doi: 10.1371/journal.pgen.1004903. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569619 Free PMC article.
-
Quantitative proteomic analysis reveals concurrent RNA-protein interactions and identifies new RNA-binding proteins in Saccharomyces cerevisiae.Genome Res. 2013 Jun;23(6):1028-38. doi: 10.1101/gr.153031.112. Epub 2013 May 1. Genome Res. 2013. PMID: 23636942 Free PMC article.
-
Oxidative stress induces coordinated remodeling of RNA-enzyme interactions.iScience. 2021 Jun 19;24(7):102753. doi: 10.1016/j.isci.2021.102753. eCollection 2021 Jul 23. iScience. 2021. PMID: 34278261 Free PMC article.
-
Regulation of histone gene transcription in yeast.Cell Mol Life Sci. 2014 Feb;71(4):599-613. doi: 10.1007/s00018-013-1443-9. Epub 2013 Aug 23. Cell Mol Life Sci. 2014. PMID: 23974242 Free PMC article. Review.
References
-
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR 2004. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329 - PubMed
-
- Avery SV 2001. Metal toxicity in yeasts and the role of oxidative stress. Adv Appl Microbiol 49: 111–142 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
