A novel universal primer-multiplex-PCR method with sequencing gel electrophoresis analysis
- PMID: 22272223
- PMCID: PMC3260127
- DOI: 10.1371/journal.pone.0022900
A novel universal primer-multiplex-PCR method with sequencing gel electrophoresis analysis
Abstract
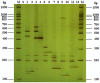
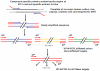
In this study, a novel universal primer-multiplex-PCR (UP-M-PCR) method adding a universal primer (UP) in the multiplex PCR reaction system was described. A universal adapter was designed in the 5'-end of each specific primer pairs which matched with the specific DNA sequences for each template and also used as the universal primer (UP). PCR products were analyzed on sequencing gel electrophoresis (SGE) which had the advantage of exhibiting extraordinary resolution. This method overcame the disadvantages rooted deeply in conventional multiplex PCR such as complex manipulation, lower sensitivity, self-inhibition and amplification disparity resulting from different primers, and it got a high specificity and had a low detection limit of 0.1 ng for single kind of crops when screening the presence of genetically modified (GM) crops in mixture samples. The novel developed multiplex PCR assay with sequencing gel electrophoresis analysis will be useful in many fields, such as verifying the GM status of a sample irrespective of the crop and GM trait and so on.
Conflict of interest statement
Figures





Similar articles
-
Single universal primer multiplex ligation-dependent probe amplification with sequencing gel electrophoresis analysis.Anal Biochem. 2013 Dec 15;443(2):243-8. doi: 10.1016/j.ab.2013.09.012. Epub 2013 Sep 16. Anal Biochem. 2013. PMID: 24050969
-
Multiplex detection of transgenic maize by microdroplet PCR combined with capillary gel electrophoresis.Acta Biochim Biophys Sin (Shanghai). 2019 May 23;51(5):535-538. doi: 10.1093/abbs/gmz011. Acta Biochim Biophys Sin (Shanghai). 2019. PMID: 30811521 No abstract available.
-
Universal primer-multiplex-polymerase chain reaction (UP-M-PCR) and capillary electrophoresis-laser-induced fluorescence analysis for the simultaneous detection of six genetically modified maize lines.J Agric Food Chem. 2011 May 25;59(10):5188-94. doi: 10.1021/jf2008088. Epub 2011 Apr 19. J Agric Food Chem. 2011. PMID: 21504143
-
Reliable detection and identification of genetically modified maize, soybean, and canola by multiplex PCR analysis.J Agric Food Chem. 2003 Sep 24;51(20):5829-34. doi: 10.1021/jf0341159. J Agric Food Chem. 2003. PMID: 13129280
-
Event-specific detection of stacked genetically modified maize Bt11 x GA21 by UP-M-PCR and real-time PCR.J Agric Food Chem. 2009 Jan 28;57(2):395-402. doi: 10.1021/jf802323m. J Agric Food Chem. 2009. PMID: 19105640
Cited by
-
Nucleic Acid Detection with Ion Concentration Polarization Microfluidic Chip for Reduced Cycle Numbers of Polymerase Chain Reaction.Micromachines (Basel). 2022 Aug 26;13(9):1394. doi: 10.3390/mi13091394. Micromachines (Basel). 2022. PMID: 36144017 Free PMC article.
-
Development of Dry and Liquid Duplex Reagent Mix-Based Polymerase Chain Reaction Assays as Novel Tools for the Rapid and Easy Quantification of Bovine Leukemia Virus (BLV) Proviral Loads.Viruses. 2024 Jun 25;16(7):1016. doi: 10.3390/v16071016. Viruses. 2024. PMID: 39066179 Free PMC article.
-
Frontiers Approaches to the Diagnosis of Thrips (Thysanoptera): How Effective Are the Molecular and Electronic Detection Platforms?Insects. 2021 Oct 9;12(10):920. doi: 10.3390/insects12100920. Insects. 2021. PMID: 34680689 Free PMC article. Review.
-
A novel universal multiplex PCR improves detection of AZFc Y-chromosome microdeletions.J Assist Reprod Genet. 2014 May;31(5):613-20. doi: 10.1007/s10815-014-0204-5. Epub 2014 Mar 11. J Assist Reprod Genet. 2014. PMID: 24615019 Free PMC article.
-
A Novel Pretreatment-Free Duplex Chamber Digital PCR Detection System for the Absolute Quantitation of GMO Samples.Int J Mol Sci. 2016 Mar 18;17(3):402. doi: 10.3390/ijms17030402. Int J Mol Sci. 2016. PMID: 26999129 Free PMC article.
References
-
- Shrestha HK, Hwu K, Wang S, Liu L, Chang M. Simultaneous detection of eight genetically modified maize lines using a combination of event- and construct-specific multiplex-PCR technique. J Agric Food Chem. 2008;56:8962–8968. - PubMed
-
- Pinar A, Bozdemir N, Kocagoz T, Alacam R. Rapid detection of bacterial atypical pneumonia agents by multiplex PCR. Cent Eur J Public Health. 2004;12:3–5. - PubMed
-
- Hess CJ, Denkers F, Ossenkoppele GJ, Waisfisz Q, McElgunn CJ, et al. Gene expression profiling of minimal residual disease in acute myeloid leukaemia by novel multiplex-PCR-based method. Leukemia. 2004;18:1981–1988. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

