Genome-wide progesterone receptor binding: cell type-specific and shared mechanisms in T47D breast cancer cells and primary leiomyoma cells
- PMID: 22272226
- PMCID: PMC3260146
- DOI: 10.1371/journal.pone.0029021
Genome-wide progesterone receptor binding: cell type-specific and shared mechanisms in T47D breast cancer cells and primary leiomyoma cells
Abstract
Background: Progesterone, via its nuclear receptor (PR), exerts an overall tumorigenic effect on both uterine fibroid (leiomyoma) and breast cancer tissues, whereas the antiprogestin RU486 inhibits growth of these tissues through an unknown mechanism. Here, we determined the interaction between common or cell-specific genome-wide binding sites of PR and mRNA expression in RU486-treated uterine leiomyoma and breast cancer cells.
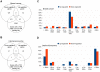
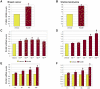
Principal findings: ChIP-sequencing revealed 31,457 and 7,034 PR-binding sites in breast cancer and uterine leiomyoma cells, respectively; 1,035 sites overlapped in both cell types. Based on the chromatin-PR interaction in both cell types, we statistically refined the consensus progesterone response element to G•ACA• • •TGT•C. We identified two striking differences between uterine leiomyoma and breast cancer cells. First, the cis-regulatory elements for HSF, TEF-1, and C/EBPα and β were statistically enriched at genomic RU486/PR-targets in uterine leiomyoma, whereas E2F, FOXO1, FOXA1, and FOXF sites were preferentially enriched in breast cancer cells. Second, 51.5% of RU486-regulated genes in breast cancer cells but only 6.6% of RU486-regulated genes in uterine leiomyoma cells contained a PR-binding site within 5 kb from their transcription start sites (TSSs), whereas 75.4% of RU486-regulated genes contained a PR-binding site farther than 50 kb from their TSSs in uterine leiomyoma cells. RU486 regulated only seven mRNAs in both cell types. Among these, adipophilin (PLIN2), a pro-differentiation gene, was induced via RU486 and PR via the same regulatory region in both cell types.
Conclusions: Our studies have identified molecular components in a RU486/PR-controlled gene network involved in the regulation of cell growth, cell migration, and extracellular matrix function. Tissue-specific and common patterns of genome-wide PR binding and gene regulation may determine the therapeutic effects of antiprogestins in uterine fibroids and breast cancer.
Conflict of interest statement
Figures


References
-
- Shyamala G. Progesterone signaling and mammary gland morphogenesis. J Mammary Gland Biol Neoplasia. 1999;4:89–104. - PubMed
-
- Mulac-Jericevic B, Mullinax RA, DeMayo FJ, Lydon JP, Conneely OM. Subgroup of reproductive functions of progesterone mediated by progesterone receptor-B isoform. Science. 2000;289:1751–1754. - PubMed
-
- Conneely OM, Mulac-Jericevic B, Lydon JP. Progesterone-dependent regulation of female reproductive activity by two distinct progesterone receptor isoforms. Steroids. 2003;68:771–778. - PubMed
-
- Bulun SE, Cheng YH, Yin P, Imir G, Utsunomiya H, et al. Progesterone resistance in endometriosis: link to failure to metabolize estradiol. Mol Cell Endocrinol. 2006;248:94–103. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

