Evolution of the primate APOBEC3A cytidine deaminase gene and identification of related coding regions
- PMID: 22272271
- PMCID: PMC3260193
- DOI: 10.1371/journal.pone.0030036
Evolution of the primate APOBEC3A cytidine deaminase gene and identification of related coding regions
Abstract
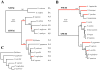
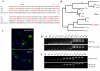
The APOBEC3 gene cluster encodes six cytidine deaminases (A3A-C, A3DE, A3F-H) with single stranded DNA (ssDNA) substrate specificity. For the moment A3A is the only enzyme that can initiate catabolism of both mitochondrial and nuclear DNA. Human A3A expression is initiated from two different methionine codons M1 or M13, both of which are in adequate but sub-optimal Kozak environments. In the present study, we have analyzed the genetic diversity among A3A genes across a wide range of 12 primates including New World monkeys, Old World monkeys and Hominids. Sequence variation was observed in exons 1-4 in all primates with up to 31% overall amino acid variation. Importantly for 3 hominids codon M1 was mutated to a threonine codon or valine codon, while for 5/12 primates strong Kozak M1 or M13 codons were found. Positive selection was apparent along a few branches which differed compared to positive selection in the carboxy-terminal of A3G that clusters with A3A among human cytidine deaminases. In the course of analyses, two novel non-functional A3A-related fragments were identified on chromosome 4 and 8 kb upstream of the A3 locus. This qualitative and quantitative variation among primate A3A genes suggest that subtle differences in function might ensue as more light is shed on this increasingly important enzyme.
Conflict of interest statement
Figures

References
-
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. - PubMed
-
- Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. - PubMed
-
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. - PubMed
-
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. - PubMed
-
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

