Molecular dynamics studies of the nucleoprotein of influenza A virus: role of the protein flexibility in RNA binding
- PMID: 22272272
- PMCID: PMC3260217
- DOI: 10.1371/journal.pone.0030038
Molecular dynamics studies of the nucleoprotein of influenza A virus: role of the protein flexibility in RNA binding
Abstract
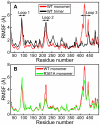
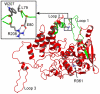
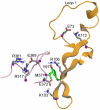
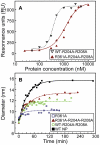
The influenza viruses contain a segmented, negative stranded RNA genome. Each RNA segment is covered by multiple copies of the nucleoprotein (NP). X-ray structures have shown that NP contains well-structured domains juxtaposed with regions of missing electron densities corresponding to loops. In this study, we tested if these flexible loops gated or promoted RNA binding and RNA-induced oligomerization of NP. We first performed molecular dynamics simulations of wt NP monomer and trimer in comparison with the R361A protein mutated in the RNA binding groove, using the H1N1 NP as the initial structure. Calculation of the root-mean-square fluctuations highlighted the presence of two flexible loops in NP trimer: loop 1 (73-90), loop 2 (200-214). In NP, loops 1 and 2 formed a 10-15 Å-wide pinch giving access to the RNA binding groove. Loop 1 was stabilized by interactions with K113 of the adjacent β-sheet 1 (91-112) that interacted with the RNA grove (linker 360-373) via multiple hydrophobic contacts. In R361A, a salt bridge formed between E80 of loop 1 and R208 of loop 2 driven by hydrophobic contacts between L79 and W207, due to a decreased flexibility of loop 2 and loop 1 unfolding. Thus, RNA could not access its binding groove in R361A; accordingly, R361A had a much lower affinity for RNA than NP. Disruption of the E80-R208 interaction in the triple mutant R361A-E80A-E81A increased its RNA binding affinity and restored its oligomerization back to wt levels in contrast with impaired levels of R361A. Our data suggest that the flexibility of loops 1 and 2 is required for RNA sampling and binding which likely involve conformational change(s) of the nucleoprotein.
Conflict of interest statement
Figures



Similar articles
-
Structure and sequence analysis of influenza A virus nucleoprotein.Sci China C Life Sci. 2009 May;52(5):439-49. doi: 10.1007/s11427-009-0064-x. Epub 2009 May 27. Sci China C Life Sci. 2009. PMID: 19471866 Review.
-
Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein.J Virol. 2009 May;83(9):4153-62. doi: 10.1128/JVI.02642-08. Epub 2009 Feb 18. J Virol. 2009. PMID: 19225007 Free PMC article.
-
Functional analysis of the influenza virus H5N1 nucleoprotein tail loop reveals amino acids that are crucial for oligomerization and ribonucleoprotein activities.J Virol. 2010 Jul;84(14):7337-45. doi: 10.1128/JVI.02474-09. Epub 2010 May 12. J Virol. 2010. PMID: 20463064 Free PMC article.
-
The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA.Nature. 2006 Dec 21;444(7122):1078-82. doi: 10.1038/nature05379. Epub 2006 Dec 6. Nature. 2006. PMID: 17151603
-
Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus.Antimicrob Agents Chemother. 2013 May;57(5):2231-42. doi: 10.1128/AAC.02335-12. Epub 2013 Mar 4. Antimicrob Agents Chemother. 2013. PMID: 23459490 Free PMC article.
Cited by
-
Monomeric nucleoprotein of influenza A virus.PLoS Pathog. 2013 Mar;9(3):e1003275. doi: 10.1371/journal.ppat.1003275. Epub 2013 Mar 28. PLoS Pathog. 2013. PMID: 23555270 Free PMC article.
-
A Universal Influenza Vaccine Can Lead to Disease Exacerbation or Viral Control Depending on Delivery Strategies.Front Immunol. 2016 Dec 26;7:641. doi: 10.3389/fimmu.2016.00641. eCollection 2016. Front Immunol. 2016. PMID: 28082980 Free PMC article.
-
The RNA-binding domain of influenzavirus non-structural protein-1 cooperatively binds to virus-specific RNA sequences in a structure-dependent manner.Nucleic Acids Res. 2013 Jan 7;41(1):434-49. doi: 10.1093/nar/gks979. Epub 2012 Oct 23. Nucleic Acids Res. 2013. PMID: 23093596 Free PMC article.
-
Molecular determinants of Ebola nucleocapsid stability from molecular dynamics simulations.J Chem Phys. 2020 Oct 21;153(15):155102. doi: 10.1063/5.0021491. J Chem Phys. 2020. PMID: 33092380 Free PMC article.
-
Comparing the decay of human wastewater-associated markers and enteric viruses in laboratory microcosms simulating estuarine waters in a temperate climatic zone using qPCR/RT-qPCR assays.Sci Total Environ. 2024 Jan 15;908:167845. doi: 10.1016/j.scitotenv.2023.167845. Epub 2023 Oct 23. Sci Total Environ. 2024. PMID: 37879463 Free PMC article.
References
-
- Elton D, Medcalf E, Bishop K, Digard P. Oligomerization of the influenza virus nucleoprotein: identification of positive and negative sequence elements. Virology. 1999;260:190–200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

