Biapenem inactivation by B2 metallo β-lactamases: energy landscape of the post-hydrolysis reactions
- PMID: 22272276
- PMCID: PMC3260057
- DOI: 10.1371/journal.pone.0030079
Biapenem inactivation by B2 metallo β-lactamases: energy landscape of the post-hydrolysis reactions
Abstract
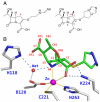
Background: The first line of defense by bacteria against β-lactam antibiotics is the expression of β-lactamases, which cleave the amide bond of the β-lactam ring. In the reaction of biapenem inactivation by B2 metallo β-lactamases (MβLs), after the β-lactam ring is opened, the carboxyl group generated by the hydrolytic process and the hydroxyethyl group (common to all carbapenems) rotate around the C5-C6 bond, assuming a new position that allows a proton transfer from the hydroxyethyl group to C2, and a nucleophilic attack on C3 by the oxygen atom of the same side-chain. This process leads to the formation of a bicyclic compound, as originally observed in the X-ray structure of the metallo β-lactamase CphA in complex with product.
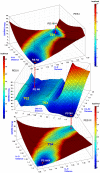
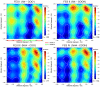
Methodology/principal findings: QM/MM and metadynamics simulations of the post-hydrolysis steps in solution and in the enzyme reveal that while the rotation of the hydroxyethyl group can occur in solution or in the enzyme active site, formation of the bicyclic compound occurs primarily in solution, after which the final product binds back to the enzyme. The calculations also suggest that the rotation and cyclization steps can occur at a rate comparable to that observed experimentally for the enzymatic inactivation of biapenem only if the hydrolysis reaction leaves the N4 nitrogen of the β-lactam ring unprotonated.
Conclusions/significance: The calculations support the existence of a common mechanism (in which ionized N4 is the leaving group) for carbapenems hydrolysis in all MβLs, and suggest a possible revision of mechanisms for B2 MβLs in which the cleavage of the β-lactam ring is associated with or immediately followed by protonation of N4. The study also indicates that the bicyclic derivative of biapenem has significant affinity for B2 MβLs, and that it may be possible to obtain clinically effective inhibitors of these enzymes by modification of this lead compound.
Conflict of interest statement
Figures






Similar articles
-
Biapenem inactivation by B2 metallo β-lactamases: energy landscape of the hydrolysis reaction.PLoS One. 2013;8(1):e55136. doi: 10.1371/journal.pone.0055136. Epub 2013 Jan 24. PLoS One. 2013. PMID: 23372827 Free PMC article.
-
Common mechanistic features among metallo-beta-lactamases: a computational study of Aeromonas hydrophila CphA enzyme.J Biol Chem. 2009 Oct 9;284(41):28164-28171. doi: 10.1074/jbc.M109.049502. Epub 2009 Aug 11. J Biol Chem. 2009. PMID: 19671702 Free PMC article.
-
A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem.J Mol Biol. 2005 Jan 28;345(4):785-95. doi: 10.1016/j.jmb.2004.10.070. J Mol Biol. 2005. PMID: 15588826
-
Metallo-β-Lactamases: Influence of the Active Site Structure on the Mechanisms of Antibiotic Resistance and Inhibition.Biochemistry (Mosc). 2021 Jan;86(Suppl 1):S24-S37. doi: 10.1134/S0006297921140030. Biochemistry (Mosc). 2021. PMID: 33827398 Review.
-
Structure, Function of Serine and Metallo-β-lactamases and their Inhibitors.Curr Protein Pept Sci. 2018;19(2):130-144. doi: 10.2174/0929866524666170724160623. Curr Protein Pept Sci. 2018. PMID: 28745223 Review.
Cited by
-
Structural Insights into Recognition of Hydrolyzed Carbapenems and Inhibitors by Subclass B3 Metallo-β-Lactamase SMB-1.Antimicrob Agents Chemother. 2016 Jun 20;60(7):4274-82. doi: 10.1128/AAC.03108-15. Print 2016 Jul. Antimicrob Agents Chemother. 2016. PMID: 27161644 Free PMC article.
-
Novel β-lactamase inhibitors: a therapeutic hope against the scourge of multidrug resistance.Front Microbiol. 2013 Dec 24;4:392. doi: 10.3389/fmicb.2013.00392. Front Microbiol. 2013. PMID: 24399995 Free PMC article.
-
Biapenem inactivation by B2 metallo β-lactamases: energy landscape of the hydrolysis reaction.PLoS One. 2013;8(1):e55136. doi: 10.1371/journal.pone.0055136. Epub 2013 Jan 24. PLoS One. 2013. PMID: 23372827 Free PMC article.
-
QM/MM molecular dynamics studies of metal binding proteins.Biomolecules. 2014 Jul 8;4(3):616-45. doi: 10.3390/biom4030616. Biomolecules. 2014. PMID: 25006697 Free PMC article. Review.
References
-
- Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. - PubMed
-
- Hall BG, Salipante SJ, Barlow M. The metallo-beta-lactamases fall into two distinct phylogenetic groups. J Mol Evol. 2003;57:249–254. - PubMed
-
- Hall BG, Barlow M. Revised Ambler classification of beta-lactamases. Journal of Antimicrobial Chemotherapy. 2005;55:1050–1051. - PubMed
-
- Frere JM, Galleni M, Bush K, Dideberg O. Is it necessary to change the classification of {beta}-lactamases? J Antimicrob Chemother. 2005;55:1051–1053. - PubMed
-
- Bebrone C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol. 2007;74:1686–1701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

