Gibberellin and auxin influence the diurnal transcription pattern of photoreceptor genes via CRY1a in tomato
- PMID: 22272283
- PMCID: PMC3260215
- DOI: 10.1371/journal.pone.0030121
Gibberellin and auxin influence the diurnal transcription pattern of photoreceptor genes via CRY1a in tomato
Abstract
Background: Plant photoreceptors, phytochromes and cryptochromes, regulate many aspects of development and growth, such as seed germination, stem elongation, seedling de-etiolation, cotyledon opening, flower induction and circadian rhythms. There are several pieces of evidence of interaction between photoreceptors and phyto-hormones in all of these physiological processes, but little is known about molecular and genetic mechanisms underlying hormone-photoreceptor crosstalk.
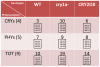
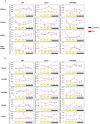
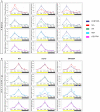
Methodology/principal findings: In this work, we investigated the molecular effects of exogenous phyto-hormones to photoreceptor gene transcripts of tomato wt, as well as transgenic and mutant lines with altered cryptochromes, by monitoring day/night transcript oscillations. GA and auxin alter the diurnal expression level of different photoreceptor genes in tomato, especially in mutants that lack a working form of cryptochrome 1a: in those mutants the expression of some (IAA) or most (GA) photoreceptor genes is down regulated by these hormones.
Conclusions/significance: Our results highlight the presence of molecular relationships among cryptochrome 1a protein, hormones, and photoreceptors' gene expression in tomato, suggesting that manipulation of cryptochromes could represent a good strategy to understand in greater depth the role of phyto-hormones in the plant photoperceptive mechanism.
Conflict of interest statement
Figures


References
-
- Sakai T, Nagashima A, Uehara Y. The ABC subfamily B auxin transporter AtABCB19 is involved in the inhibitory effects of N-1-Naphthyphthalamic acid on the phototropic and gravitropic responses of Arabidopsis hypocotyls. Plant and Cell Physiology. 2008;49:1250–1255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

