The translation elongation factor eEF-1Bβ1 is involved in cell wall biosynthesis and plant development in Arabidopsis thaliana
- PMID: 22272350
- PMCID: PMC3260303
- DOI: 10.1371/journal.pone.0030425
The translation elongation factor eEF-1Bβ1 is involved in cell wall biosynthesis and plant development in Arabidopsis thaliana
Abstract
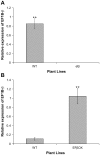
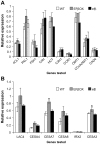
The eukaryotic translation elongation factor eEF-1Bβ1 (EF1Bβ) is a guanine nucleotide exchange factor that plays an important role in translation elongation. In this study, we show that the EF1Bβ protein is localized in the plasma membrane and cytoplasm, and that the transcripts should be expressed in most tissue types in seedlings. Sectioning of the inflorescence stem revealed that EF1Bβ predominantly localizes to the xylem vessels and in the interfascicular cambium. EF1Bβ gene silencing in efβ caused a dwarf phenotype with 38% and 20% reduction in total lignin and crystalline cellulose, respectively. This loss-of-function mutant also had a lower S/G lignin monomer ratio relative to wild type plants, but no changes were detected in a gain-of-function mutant transformed with the EF1Bβ gene. Histochemical analysis showed a reduced vascular apparatus, including smaller xylem vessels in the inflorescence stem of the loss-of-function mutant. Over-expression of EF1Bβ in an eli1 mutant background restored a WT phenotype and abolished ectopic lignin deposition as well as cell expansion defects in the mutant. Taken together, these data strongly suggest a role for EF1Bβ in plant development and cell wall formation in Arabidopsis.
Conflict of interest statement
Figures







Similar articles
-
DIMINUTO 1 affects the lignin profile and secondary cell wall formation in Arabidopsis.Planta. 2012 Mar;235(3):485-98. doi: 10.1007/s00425-011-1519-4. Epub 2011 Sep 27. Planta. 2012. PMID: 21947665
-
The Arabidopsis Domain of Unknown Function 1218 (DUF1218) Containing Proteins, MODIFYING WALL LIGNIN-1 and 2 (At1g31720/MWL-1 and At4g19370/MWL-2) Function Redundantly to Alter Secondary Cell Wall Lignin Content.PLoS One. 2016 Mar 1;11(3):e0150254. doi: 10.1371/journal.pone.0150254. eCollection 2016. PLoS One. 2016. PMID: 26930070 Free PMC article.
-
A R2R3-MYB transcription factor that is specifically expressed in cotton (Gossypium hirsutum) fibers affects secondary cell wall biosynthesis and deposition in transgenic Arabidopsis.Physiol Plant. 2015 Jul;154(3):420-32. doi: 10.1111/ppl.12317. Epub 2015 Jan 26. Physiol Plant. 2015. PMID: 25534543
-
The Arabidopsis wood model-the case for the inflorescence stem.Plant Sci. 2013 Sep;210:193-205. doi: 10.1016/j.plantsci.2013.05.007. Epub 2013 May 21. Plant Sci. 2013. PMID: 23849126 Review.
-
A review of xylan and lignin biosynthesis: foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes.Crit Rev Biochem Mol Biol. 2014 May-Jun;49(3):212-41. doi: 10.3109/10409238.2014.889651. Epub 2014 Feb 24. Crit Rev Biochem Mol Biol. 2014. PMID: 24564339 Review.
Cited by
-
UVSSA, UBP12, and RDO2/TFIIS Contribute to Arabidopsis UV Tolerance.Front Plant Sci. 2019 Apr 24;10:516. doi: 10.3389/fpls.2019.00516. eCollection 2019. Front Plant Sci. 2019. PMID: 31105721 Free PMC article.
-
Transcriptome characteristics and six alternative expressed genes positively correlated with the phase transition of annual cambial activities in Chinese Fir (Cunninghamia lanceolata (Lamb.) Hook).PLoS One. 2013 Aug 12;8(8):e71562. doi: 10.1371/journal.pone.0071562. eCollection 2013. PLoS One. 2013. PMID: 23951189 Free PMC article.
-
QTL Identification for Stem Fiber, Strength and Rot Resistance in a DH Population from an Alien Introgression of Brassica napus.Plants (Basel). 2022 Jan 29;11(3):373. doi: 10.3390/plants11030373. Plants (Basel). 2022. PMID: 35161354 Free PMC article.
-
The Arabidopsis cytosolic proteome: the metabolic heart of the cell.Front Plant Sci. 2014 Feb 5;5:21. doi: 10.3389/fpls.2014.00021. eCollection 2014. Front Plant Sci. 2014. PMID: 24550929 Free PMC article. Review.
-
Role of Arabidopsis ABF1/3/4 during det1 germination in salt and osmotic stress conditions.Plant Mol Biol. 2018 May;97(1-2):149-163. doi: 10.1007/s11103-018-0729-6. Epub 2018 Apr 21. Plant Mol Biol. 2018. PMID: 29680877
References
-
- Mathews MB, Sonenberg N, Hershey JWB. Origins and principles of translational control. In: Sonenberg N, Hershey J, Mathews M, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 1–31.
-
- Merrick WC, Nyborg J. The protein synthesis elongation cycle. In: Sonenberg N, Hershey J, Mathews M, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 89–126.
-
- Hiraga K, Suzuki K, Tsuchiya E, Miyakawa T. Cloning and characterization of the elongation factor EF-1β homologue of Saccharomyces cerevisiae; EF-1β is essential for growth. FEBS Lett. 1993;316:165–169. - PubMed
-
- Olarewaju O, Ortiz PA, Chowdhury WQ, Chatterjee I, Kinzy TG. The translation elongation factor eEF1B plays a role in the oxidative stress response pathway. RNA Biol. 2004;1:89–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

