Expression profiling reveals Spot 42 small RNA as a key regulator in the central metabolism of Aliivibrio salmonicida
- PMID: 22272603
- PMCID: PMC3295665
- DOI: 10.1186/1471-2164-13-37
Expression profiling reveals Spot 42 small RNA as a key regulator in the central metabolism of Aliivibrio salmonicida
Abstract
Background: Spot 42 was discovered in Escherichia coli nearly 40 years ago as an abundant, small and unstable RNA. Its biological role has remained obscure until recently, and is today implicated in having broader roles in the central and secondary metabolism. Spot 42 is encoded by the spf gene. The gene is ubiquitous in the Vibrionaceae family of gamma-proteobacteria. One member of this family, Aliivibrio salmonicida, causes cold-water vibriosis in farmed Atlantic salmon. Its genome encodes Spot 42 with 84% identity to E. coli Spot 42.
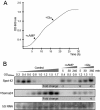
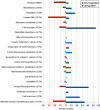
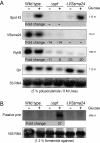
Results: We generated a A. salmonicida spf deletion mutant. We then used microarray and Northern blot analyses to monitor global effects on the transcriptome in order to provide insights into the biological roles of Spot 42 in this bacterium. In the presence of glucose, we found a surprisingly large number of ≥ 2X differentially expressed genes, and several major cellular processes were affected. A gene encoding a pirin-like protein showed an on/off expression pattern in the presence/absence of Spot 42, which suggests that Spot 42 plays a key regulatory role in the central metabolism by regulating the switch between fermentation and respiration. Interestingly, we discovered an sRNA named VSsrna24, which is encoded immediately downstream of spf. This new sRNA has an expression pattern opposite to that of Spot 42, and its expression is repressed by glucose.
Conclusions: We hypothesize that Spot 42 plays a key role in the central metabolism, in part by regulating the pyruvat dehydrogenase enzyme complex via pirin.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

