Genetic diversity of human RNase 8
- PMID: 22272736
- PMCID: PMC3295680
- DOI: 10.1186/1471-2164-13-40
Genetic diversity of human RNase 8
Abstract
Background: Ribonuclease 8 is a member of the RNase A family of secretory ribonucleases; orthologs of this gene have been found only in primate genomes. RNase 8 is a divergent paralog of RNase 7, which is lysine-enriched, highly conserved, has prominent antimicrobial activity, and is expressed in both normal and diseased skin; in contrast, the physiologic function of RNase 8 remains uncertain. Here, we examine the genetic diversity of human RNase 8, a subject of significant interest given the existence of functional pseudogenes (coding sequences that are otherwise intact but with mutations in elements crucial for ribonucleolytic activity) in non-human primate genomes.
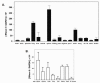
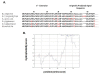
Results: RNase 8 expression was detected in adult human lung, spleen and testis tissue by quantitative reverse-transcription PCR. Only two single-nucleotide polymorphisms and four unique alleles were identified within the RNase 8 coding sequence; nucleotide sequence diversity (π = 0.00122 ± 0.00009 per site) was unremarkable for a human nuclear gene. We isolated transcripts encoding RNase 8 via rapid amplification of cDNA ends (RACE) and RT-PCR which included a distal potential translational start site followed by sequence encoding an additional 30 amino acids that are conserved in the genomes of several higher primates. The distal translational start site is functional and promotes RNase 8 synthesis in transfected COS-7 cells.
Conclusions: These results suggest that RNase 8 may diverge considerably from typical RNase A family ribonucleases and may likewise exhibit unique function. This finding prompts a reconsideration of what we have previously termed functional pseudogenes, as RNase 8 may be responding to constraints that promote significant functional divergence from the canonical structure and enzymatic activity characteristic of the RNase A family.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

