The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA
- PMID: 22274953
- PMCID: PMC3285931
- DOI: 10.1261/rna.030841.111
The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA
Abstract
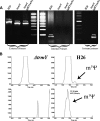
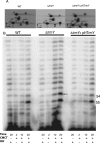
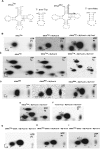
The methylation of pseudouridine (Ψ) at position 54 of tRNA, producing m(1)Ψ, is a hallmark of many archaeal species, but the specific methylase involved in the formation of this modification had yet to be characterized. A comparative genomics analysis had previously identified COG1901 (DUF358), part of the SPOUT superfamily, as a candidate for this missing methylase family. To test this prediction, the COG1901 encoding gene, HVO_1989, was deleted from the Haloferax volcanii genome. Analyses of modified base contents indicated that while m(1)Ψ was present in tRNA extracted from the wild-type strain, it was absent from tRNA extracted from the mutant strain. Expression of the gene encoding COG1901 from Halobacterium sp. NRC-1, VNG1980C, complemented the m(1)Ψ minus phenotype of the ΔHVO_1989 strain. This in vivo validation was extended with in vitro tests. Using the COG1901 recombinant enzyme from Methanocaldococcus jannaschii (Mj1640), purified enzyme Pus10 from M. jannaschii and full-size tRNA transcripts or TΨ-arm (17-mer) fragments as substrates, the sequential pathway of m(1)Ψ54 formation in Archaea was reconstituted. The methylation reaction is AdoMet dependent. The efficiency of the methylase reaction depended on the identity of the residue at position 55 of the TΨ-loop. The presence of Ψ55 allowed the efficient conversion of Ψ54 to m(1)Ψ54, whereas in the presence of C55, the reaction was rather inefficient and no methylation reaction occurred if a purine was present at this position. These results led to renaming the Archaeal COG1901 members as TrmY proteins.
Figures

Similar articles
-
Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs.RNA. 2012 Mar;18(3):412-20. doi: 10.1261/rna.028498.111. Epub 2012 Jan 24. RNA. 2012. PMID: 22274954 Free PMC article.
-
Pseudouridine formation in archaeal RNAs: The case of Haloferax volcanii.RNA. 2011 Jul;17(7):1367-80. doi: 10.1261/rna.2712811. Epub 2011 May 31. RNA. 2011. PMID: 21628430 Free PMC article.
-
Role of forefinger and thumb loops in production of Ψ54 and Ψ55 in tRNAs by archaeal Pus10.RNA. 2013 Sep;19(9):1279-94. doi: 10.1261/rna.039230.113. Epub 2013 Jul 29. RNA. 2013. PMID: 23898217 Free PMC article.
-
Pseudouridines and pseudouridine synthases of the ribosome.Cold Spring Harb Symp Quant Biol. 2001;66:147-59. doi: 10.1101/sqb.2001.66.147. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762017 Review.
-
Transfer RNA processing in archaea: unusual pathways and enzymes.FEBS Lett. 2010 Jan 21;584(2):303-9. doi: 10.1016/j.febslet.2009.10.067. FEBS Lett. 2010. PMID: 19878676 Free PMC article. Review.
Cited by
-
The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced-fit process.J Biol Chem. 2013 Aug 30;288(35):25562-25574. doi: 10.1074/jbc.M113.485128. Epub 2013 Jul 18. J Biol Chem. 2013. PMID: 23867454 Free PMC article.
-
Tied up in knots: Untangling substrate recognition by the SPOUT methyltransferases.J Biol Chem. 2022 Oct;298(10):102393. doi: 10.1016/j.jbc.2022.102393. Epub 2022 Aug 18. J Biol Chem. 2022. PMID: 35988649 Free PMC article. Review.
-
Transfer RNA Modification Enzymes from Thermophiles and Their Modified Nucleosides in tRNA.Microorganisms. 2018 Oct 20;6(4):110. doi: 10.3390/microorganisms6040110. Microorganisms. 2018. PMID: 30347855 Free PMC article. Review.
-
Reciprocal amplification of caspase-3 activity by nuclear export of a putative human RNA-modifying protein, PUS10 during TRAIL-induced apoptosis.Cell Death Dis. 2017 Oct 5;8(10):e3093. doi: 10.1038/cddis.2017.476. Cell Death Dis. 2017. PMID: 28981101 Free PMC article.
-
Naturally occurring modified ribonucleosides.Wiley Interdiscip Rev RNA. 2020 Sep;11(5):e1595. doi: 10.1002/wrna.1595. Epub 2020 Apr 16. Wiley Interdiscip Rev RNA. 2020. PMID: 32301288 Free PMC article. Review.
References
-
- Bakin AV, Ofengand J 1998. Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol Biol 77: 297–309 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
