Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs
- PMID: 22274954
- PMCID: PMC3285930
- DOI: 10.1261/rna.028498.111
Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs
Abstract
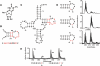
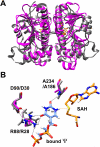
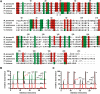
tRNAs from all three kingdoms of life contain a variety of modified nucleotides required for their stability, proper folding, and accurate decoding. One prominent example is the eponymous ribothymidine (rT) modification at position 54 in the T-arm of eukaryotic and bacterial tRNAs. In contrast, in most archaea this position is occupied by another hypermodified nucleotide: the isosteric N1-methylated pseudouridine. While the enzyme catalyzing pseudouridine formation at this position is known, the pseudouridine N1-specific methyltransferase responsible for this modification has not yet been experimentally identified. Here, we present biochemical and genetic evidence that the two homologous proteins, Mja_1640 (COG 1901, Pfam DUF358) and Hvo_1989 (Pfam DUF358) from Methanocaldococcus jannaschii and Haloferax volcanii, respectively, are representatives of the methyltransferase responsible for this modification. However, the in-frame deletion of the pseudouridine N1-methyltransferase gene in H. volcanii did not result in a discernable phenotype in line with similar observations for knockouts of other T-arm methylating enzymes.
Figures

Similar articles
-
The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA.RNA. 2012 Mar;18(3):421-33. doi: 10.1261/rna.030841.111. Epub 2012 Jan 24. RNA. 2012. PMID: 22274953 Free PMC article.
-
Pseudouridine formation in archaeal RNAs: The case of Haloferax volcanii.RNA. 2011 Jul;17(7):1367-80. doi: 10.1261/rna.2712811. Epub 2011 May 31. RNA. 2011. PMID: 21628430 Free PMC article.
-
The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase.Nucleic Acids Res. 2010 Apr;38(7):2387-98. doi: 10.1093/nar/gkp1189. Epub 2010 Jan 4. Nucleic Acids Res. 2010. PMID: 20047967 Free PMC article.
-
Pseudouridines and pseudouridine synthases of the ribosome.Cold Spring Harb Symp Quant Biol. 2001;66:147-59. doi: 10.1101/sqb.2001.66.147. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762017 Review.
-
Identification and characterization of archaeal and fungal tRNA methyltransferases.Methods Enzymol. 2007;425:185-209. doi: 10.1016/S0076-6879(07)25008-6. Methods Enzymol. 2007. PMID: 17673084 Review.
Cited by
-
Identification of the 3-amino-3-carboxypropyl (acp) transferase enzyme responsible for acp3U formation at position 47 in Escherichia coli tRNAs.Nucleic Acids Res. 2020 Feb 20;48(3):1435-1450. doi: 10.1093/nar/gkz1191. Nucleic Acids Res. 2020. PMID: 31863583 Free PMC article.
-
tRNA Modification Profiles and Codon-Decoding Strategies in Methanocaldococcus jannaschii.J Bacteriol. 2019 Apr 9;201(9):e00690-18. doi: 10.1128/JB.00690-18. Print 2019 May 1. J Bacteriol. 2019. PMID: 30745370 Free PMC article.
-
The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced-fit process.J Biol Chem. 2013 Aug 30;288(35):25562-25574. doi: 10.1074/jbc.M113.485128. Epub 2013 Jul 18. J Biol Chem. 2013. PMID: 23867454 Free PMC article.
-
Pseudouridine: still mysterious, but never a fake (uridine)!RNA Biol. 2014;11(12):1540-54. doi: 10.4161/15476286.2014.992278. RNA Biol. 2014. PMID: 25616362 Free PMC article. Review.
-
Post-Transcriptional Modifications of Conserved Nucleotides in the T-Loop of tRNA: A Tale of Functional Convergent Evolution.Genes (Basel). 2021 Jan 22;12(2):140. doi: 10.3390/genes12020140. Genes (Basel). 2021. PMID: 33499018 Free PMC article. Review.
References
-
- Bahr U, Aygün H, Karas M 2009. Sequencing of single and double stranded RNA oligonucleotides by acid hydrolysis and MALDI mass spectrometry. Anal Chem 81: 3173–3179 - PubMed
-
- Blobstein SH, Grunberger D, Weinstein IB, Nakanishi K 1973. Isolation and structure determination of the fluorescent base from bovine liver phenylalanine transfer ribonucleic acid. Biochemistry 12: 188–193 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
