Identification of genes required for Cf-dependent hypersensitive cell death by combined proteomic and RNA interfering analyses
- PMID: 22275387
- PMCID: PMC3346213
- DOI: 10.1093/jxb/err397
Identification of genes required for Cf-dependent hypersensitive cell death by combined proteomic and RNA interfering analyses
Abstract
Identification of hypersensitive cell death (HCD) regulators is essential to dissect the molecular mechanisms underlying plant disease resistance. In this study, combined proteomic and RNA interfering (RNAi) analyses were employed to identify genes required for the HCD conferred by the tomato resistance gene Cf-4 and the Cladosporium fulvum avirulence gene Avr4. Forty-nine proteins differentially expressed in the tomato seedlings mounting and those not mounting Cf-4/Avr4-dependent HCD were identified through proteomic analysis. Among them were a variety of defence-related proteins including a cysteine protease, Pip1, an operative target of another C. fulvum effector, Avr2. Additionally, glutathione-mediated antioxidation is a major response to Cf-4/Avr4-dependent HCD. Functional analysis through tobacco rattle virus-induced gene silencing and transient RNAi assays of the chosen 16 differentially expressed proteins revealed that seven genes, which encode Pip1 homologue NbPip1, a SIPK type MAP kinase Nbf4, an asparagine synthetase NbAsn, a trypsin inhibitor LeMir-like protein NbMir, a small GTP-binding protein, a late embryogenesis-like protein, and an ASR4-like protein, were required for Cf-4/Avr4-dependent HCD. Furthermore, the former four genes were essential for Cf-9/Avr9-dependent HCD; NbPip1, NbAsn, and NbMir, but not Nbf4, affected a nonadaptive bacterial pathogen Xanthomonas oryzae pv. oryzae-induced HCD in Nicotiana benthamiana. These data demonstrate that Pip1 and LeMir may play a general role in HCD and plant immunity and that the application of combined proteomic and RNA interfering analyses is an efficient strategy to identify genes required for HCD, disease resistance, and probably other biological processes in plants.
Figures








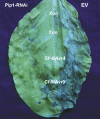
Similar articles
-
Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato.Plant Cell. 2005 Jan;17(1):295-310. doi: 10.1105/tpc.104.026013. Epub 2004 Dec 14. Plant Cell. 2005. PMID: 15598806 Free PMC article.
-
Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance.Plant J. 2010 Apr;62(2):224-39. doi: 10.1111/j.1365-313X.2010.04136.x. Epub 2010 Jan 20. Plant J. 2010. PMID: 20088897
-
Bioengineering secreted proteases converts divergent Rcr3 orthologs and paralogs into extracellular immune co-receptors.Plant Cell. 2024 Sep 3;36(9):3260-3276. doi: 10.1093/plcell/koae183. Plant Cell. 2024. PMID: 38923940 Free PMC article.
-
Molecular and biochemical basis of the interaction between tomato and its fungal pathogen Cladosporium fulvum.Antonie Van Leeuwenhoek. 1997 Feb;71(1-2):137-41. doi: 10.1023/a:1000102509556. Antonie Van Leeuwenhoek. 1997. PMID: 9049025 Review.
-
Recognitional specificity and evolution in the tomato-Cladosporium fulvum pathosystem.Mol Plant Microbe Interact. 2009 Oct;22(10):1191-202. doi: 10.1094/MPMI-22-10-1191. Mol Plant Microbe Interact. 2009. PMID: 19737093 Review.
Cited by
-
Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of Cf receptor-like proteins involved in pathogen resistance of tomato.Plant Physiol. 2012 Aug;159(4):1819-33. doi: 10.1104/pp.112.196741. Epub 2012 May 30. Plant Physiol. 2012. PMID: 22649272 Free PMC article.
-
Chaperones of the endoplasmic reticulum are required for Ve1-mediated resistance to Verticillium.Mol Plant Pathol. 2014 Jan;15(1):109-17. doi: 10.1111/mpp.12071. Epub 2013 Sep 10. Mol Plant Pathol. 2014. PMID: 24015989 Free PMC article.
-
Comparative proteomic analysis reveals the cross-talk between the responses induced by H2O2 and by long-term rice black-streaked dwarf virus infection in rice.PLoS One. 2013 Nov 27;8(11):e81640. doi: 10.1371/journal.pone.0081640. eCollection 2013. PLoS One. 2013. PMID: 24312331 Free PMC article.
-
Indispensable Role of Proteases in Plant Innate Immunity.Int J Mol Sci. 2018 Feb 23;19(2):629. doi: 10.3390/ijms19020629. Int J Mol Sci. 2018. PMID: 29473858 Free PMC article. Review.
-
The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire.Plant Biotechnol J. 2018 May;16(5):1068-1084. doi: 10.1111/pbi.12852. Epub 2017 Dec 17. Plant Biotechnol J. 2018. PMID: 29055088 Free PMC article.
References
-
- Blatt MR, Grabov A, Brearley J, Hammond-Kosack KE, Jones JDG. K+ channels of Cf-9 transgenic tobacco guard cells as targets for Cladosporium fulvum Avr9 elicitor-dependent signal transduction. The Plant Journal. 1999;19:453–462. - PubMed
-
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Applications and advantages of virus-induced gene silencing for gene function studies in plants. The Plant Journal. 2004;39:734–746. - PubMed
-
- Cai X, Takken FLW, Joosten MHAJ, De Wit PJGM. Specific recognition of AVR4 and AVR9 results in distinct patterns of hypersensitive cell death in tomato, but similar patterns of defence-related gene expression. Molecular Plant Pathology. 2001;2:77–86. - PubMed
-
- Cai XZ, Wang CC, Xu YP, Xu QF, Zheng Z, Zhou XP. Efficient gene silencing induction in tomato by a viral satellite DNA vector. Virus Research. 2007;125:169–175. - PubMed

