Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides
- PMID: 22275502
- PMCID: PMC3262234
- DOI: 10.1128/mBio.00279-11
Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides
Abstract
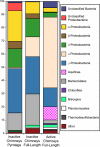
Hydrothermal chimneys are a globally dispersed habitat on the seafloor associated with mid-ocean ridge (MOR) spreading centers. Active, hot, venting sulfide structures from MORs have been examined for microbial diversity and ecology since their discovery in the mid-1970s, and recent work has also begun to explore the microbiology of inactive sulfides--structures that persist for decades to millennia and form moderate to massive deposits at and below the seafloor. Here we used tag pyrosequencing of the V6 region of the 16S rRNA and full-length 16S rRNA sequencing on inactive hydrothermal sulfide chimney samples from 9°N on the East Pacific Rise to learn their bacterial composition, metabolic potential, and succession from venting to nonventing (inactive) regimes. Alpha-, beta-, delta-, and gammaproteobacteria and members of the phylum Bacteroidetes dominate all inactive sulfides. Greater than 26% of the V6 tags obtained are closely related to lineages involved in sulfur, nitrogen, iron, and methane cycling. Epsilonproteobacteria represent <4% of the V6 tags recovered from inactive sulfides and 15% of the full-length clones, despite their high abundance in active chimneys. Members of the phylum Aquificae, which are common in active vents, were absent from both the V6 tags and full-length 16S rRNA data sets. In both analyses, the proportions of alphaproteobacteria, betaproteobacteria, and members of the phylum Bacteroidetes were greater than those found on active hydrothermal sulfides. These shifts in bacterial population structure on inactive chimneys reveal ecological succession following cessation of venting and also imply a potential shift in microbial activity and metabolic guilds on hydrothermal sulfides, the dominant biome that results from seafloor venting.
Figures




References
-
- Hannington MD, Jonasson IR, Herzig PM, Petersen S. 1995. Physical and chemical processes of seafloor mineralization at mid-ocean ridges, p. 115–157 In Humphris SE, Zierenberg RA, Mullineaux LS, Thomson RE, Seafloor hydrothermal systems: physical, chemical, biological and geological interactions. American Geophysical Union, Washington, DC
-
- Hoek J, Banta A, Hubler F, Reysenbach AL. 2003. Microbial diversity of a sulphide spire located in the Edmond deep-sea hydrothermal vent field on the central Indian ridge. Geobiology 1:119–127
-
- Kormas KA, Tivey MK, Von Damm K, Teske A. 2006. Bacterial and archaeal phylotypes associated with distinct mineralogical layers of a white smoker spire from a deep-sea hydrothermal vent site (nine degrees N, East Pacific rise). Environ. Microbiol. 8:909–920 - PubMed
-
- Nakagawa S, et al. 2005. Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: impacts of subseafloor phase-separation. FEMS Microbiol. Ecol. 54:141–155 - PubMed
-
- Nakagawa S, et al. 2005. Distribution, phylogenetic diversity and physiological characteristics of epsilon-proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7:1619–1632 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
