Molecular evolution in nonrecombining regions of the Drosophila melanogaster genome
- PMID: 22275518
- PMCID: PMC3318434
- DOI: 10.1093/gbe/evs010
Molecular evolution in nonrecombining regions of the Drosophila melanogaster genome
Abstract
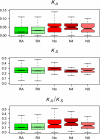
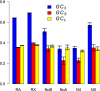
We study the evolutionary effects of reduced recombination on the Drosophila melanogaster genome, analyzing more than 200 new genes that lack crossing-over and employing a novel orthology search among species of the melanogaster subgroup. These genes are located in the heterochromatin of chromosomes other than the dot (fourth) chromosome. Noncrossover regions of the genome all exhibited an elevated level of evolutionary divergence from D. yakuba at nonsynonymous sites, lower codon usage bias, lower GC content in coding and noncoding regions, and longer introns. Levels of gene expression are similar for genes in regions with and without crossing-over, which rules out the possibility that the reduced level of adaptation that we detect is caused by relaxed selection due to lower levels of gene expression in the heterochromatin. The patterns observed are consistent with a reduction in the efficacy of selection in all regions of the genome of D. melanogaster that lack crossing-over, as a result of the effects of enhanced Hill-Robertson interference. However, we also detected differences among nonrecombining locations: The X chromosome seems to exhibit the weakest effects, whereas the fourth chromosome and the heterochromatic genes on the autosomes located most proximal to the centromere showed the largest effects. However, signatures of selection on both nonsynonymous mutations and on codon usage persist in all heterochromatic regions.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

