HIV-1 polymerase inhibition by nucleoside analogs: cellular- and kinetic parameters of efficacy, susceptibility and resistance selection
- PMID: 22275860
- PMCID: PMC3261923
- DOI: 10.1371/journal.pcbi.1002359
HIV-1 polymerase inhibition by nucleoside analogs: cellular- and kinetic parameters of efficacy, susceptibility and resistance selection
Abstract
Nucleoside analogs (NAs) are used to treat numerous viral infections and cancer. They compete with endogenous nucleotides (dNTP/NTP) for incorporation into nascent DNA/RNA and inhibit replication by preventing subsequent primer extension. To date, an integrated mathematical model that could allow the analysis of their mechanism of action, of the various resistance mechanisms, and their effect on viral fitness is still lacking. We present the first mechanistic mathematical model of polymerase inhibition by NAs that takes into account the reversibility of polymerase inhibition. Analytical solutions for the model point out the cellular- and kinetic aspects of inhibition. Our model correctly predicts for HIV-1 that resistance against nucleoside analog reverse transcriptase inhibitors (NRTIs) can be conferred by decreasing their incorporation rate, increasing their excision rate, or decreasing their affinity for the polymerase enzyme. For all analyzed NRTIs and their combinations, model-predicted macroscopic parameters (efficacy, fitness and toxicity) were consistent with observations. NRTI efficacy was found to greatly vary between distinct target cells. Surprisingly, target cells with low dNTP/NTP levels may not confer hyper-susceptibility to inhibition, whereas cells with high dNTP/NTP contents are likely to confer natural resistance. Our model also allows quantification of the selective advantage of mutations by integrating their effects on viral fitness and drug susceptibility. For zidovudine triphosphate (AZT-TP), we predict that this selective advantage, as well as the minimal concentration required to select thymidine-associated mutations (TAMs) are highly cell-dependent. The developed model allows studying various resistance mechanisms, inherent fitness effects, selection forces and epistasis based on microscopic kinetic data. It can readily be embedded in extended models of the complete HIV-1 reverse transcription process, or analogous processes in other viruses and help to guide drug development and improve our understanding of the mechanisms of resistance development during treatment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
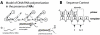
 in the model correspond to the condition in which
in the model correspond to the condition in which  nucleosides have been attached and state
nucleosides have been attached and state  corresponds to full-length product, after which the enzyme dissociates from the template/primer. States
corresponds to full-length product, after which the enzyme dissociates from the template/primer. States  correspond to the condition, in which a DNA-chain consisting of
correspond to the condition, in which a DNA-chain consisting of  natural nucleosides has been produced, but where the last
natural nucleosides has been produced, but where the last  nucleoside in the chain is a chain-terminating
nucleoside in the chain is a chain-terminating  . At each state
. At each state  , the nascent DNA-chain can either be shortened (pyrophosphorolysis
, the nascent DNA-chain can either be shortened (pyrophosphorolysis  ), -prolonged with a nucleoside (polymerase reaction
), -prolonged with a nucleoside (polymerase reaction  ) or -terminated by a nucleoside analog (reaction
) or -terminated by a nucleoside analog (reaction  ). If the chain has been terminated (state
). If the chain has been terminated (state  ), it can get released with rate
), it can get released with rate  (excision reaction) to produce a chain of length
(excision reaction) to produce a chain of length  . B: Sequence context. The reaction rates
. B: Sequence context. The reaction rates  ,
,  ,
,  and
and  depend on the nucleoside sequence of the template. In the illustration, the next incoming nucleoside could be either a thymidine or a thymidine-analog (corresponding to position
depend on the nucleoside sequence of the template. In the illustration, the next incoming nucleoside could be either a thymidine or a thymidine-analog (corresponding to position  in the template sequence). Therefore,
in the template sequence). Therefore,  and
and  would refer to thymidine- and thymidine-analog incorporation. The pyrophosphorolysis reaction, on the other hand, would refer to cytosine removal (position
would refer to thymidine- and thymidine-analog incorporation. The pyrophosphorolysis reaction, on the other hand, would refer to cytosine removal (position  in the primer sequence).
in the primer sequence).
 ddATP. The dotted red- and green lines (upward and downward pointing triangles) show the time required for polymerization in the ‘K65R’ mutant RT enzyme in the presence- and absence of
ddATP. The dotted red- and green lines (upward and downward pointing triangles) show the time required for polymerization in the ‘K65R’ mutant RT enzyme in the presence- and absence of  ddATP. Kinetic parameters are presented in Table 1 and Table S1, S2 (supplementary material) for the wild type and the ‘K65R’ mutant. B: Single nucleoside incorporation time
ddATP. Kinetic parameters are presented in Table 1 and Table S1, S2 (supplementary material) for the wild type and the ‘K65R’ mutant. B: Single nucleoside incorporation time  in the absence of ddATP in the wildtype and the ‘K65R’ mutant (solid black and dashed green lines respectively) and in the presence of ddATP in the wild type enzyme (dashed blue line) and in the mutant enzyme (dotted red line), calculated using equation eq. (8).
in the absence of ddATP in the wildtype and the ‘K65R’ mutant (solid black and dashed green lines respectively) and in the presence of ddATP in the wild type enzyme (dashed blue line) and in the mutant enzyme (dotted red line), calculated using equation eq. (8).
 T-cells (solid line) and the impact of a 100-fold variation of the the intracellular nucleoside concentrations (dotted line). The illustration was generated by evaluating eq. (19) and typical parameters for DNA-dependent polymerization during HIV-1 reverse transcription and its inhibition by ddATP (all parameters are indicated in Table 1 and Table S1, supplementary material). The corresponding
T-cells (solid line) and the impact of a 100-fold variation of the the intracellular nucleoside concentrations (dotted line). The illustration was generated by evaluating eq. (19) and typical parameters for DNA-dependent polymerization during HIV-1 reverse transcription and its inhibition by ddATP (all parameters are indicated in Table 1 and Table S1, supplementary material). The corresponding  is depicted by a green filled circle. B: Molecular mechanisms of drug resistance and hyper-susceptibility (dashed lines). Impact of (i) selective attrition of inhibitor incorporation
is depicted by a green filled circle. B: Molecular mechanisms of drug resistance and hyper-susceptibility (dashed lines). Impact of (i) selective attrition of inhibitor incorporation  and (ii) selective attrition of inhibitor binding to the primer-template
and (ii) selective attrition of inhibitor binding to the primer-template  on drug susceptibility. Hypersusceptibility is conferred by the opposite change in the indicated parameters. In order to generate the dashed lines, the respective parameters have been increased/decreased by a factor of 100.
on drug susceptibility. Hypersusceptibility is conferred by the opposite change in the indicated parameters. In order to generate the dashed lines, the respective parameters have been increased/decreased by a factor of 100.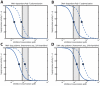
 T-cells from , converted to units
T-cells from , converted to units  by assuming a cell volume of
by assuming a cell volume of  for resting
for resting  T-cells . All utilized parameters are indicated in Tables 1, S1, S2, S3 (supplementary material).
T-cells . All utilized parameters are indicated in Tables 1, S1, S2, S3 (supplementary material).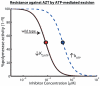
 via an ATP-mediated mechanism (see eq. (21)). We calculated residual DNA-dependent polymerization of a Poly-T sequence in unstimulated
via an ATP-mediated mechanism (see eq. (21)). We calculated residual DNA-dependent polymerization of a Poly-T sequence in unstimulated  T-cells using eq. (19) with parameters from Tables 1, S1 and S3 (supplementary material). The solid black line shows residual DNA polymerization
T-cells using eq. (19) with parameters from Tables 1, S1 and S3 (supplementary material). The solid black line shows residual DNA polymerization  in the wild type virus, whereas the dotted red line and the dashed blue line refer to residual polymerization if
in the wild type virus, whereas the dotted red line and the dashed blue line refer to residual polymerization if  and
and  were decreased- and increased 100-fold respectively.
were decreased- and increased 100-fold respectively.
 cells, blue = unstimulated
cells, blue = unstimulated  cells, red = macrophages) indicate the selection parameter
cells, red = macrophages) indicate the selection parameter  , defined in eq. (4), for different levels of intracellular ATZ-TP during RNA- and DNA dependent polymerization (Panels A & B) of a random sequence of length 500 with 25% respective dNTP content. The light grey area indicates the in vivo concentrations range of AZT in purified circulating
, defined in eq. (4), for different levels of intracellular ATZ-TP during RNA- and DNA dependent polymerization (Panels A & B) of a random sequence of length 500 with 25% respective dNTP content. The light grey area indicates the in vivo concentrations range of AZT in purified circulating  T-cells from , converted to units
T-cells from , converted to units  by assuming a cell volume of
by assuming a cell volume of  for resting
for resting  T-cells . The dashed horizontal line indicates the threshold for resistance selection, i.e.
T-cells . The dashed horizontal line indicates the threshold for resistance selection, i.e.  . All utilized parameters are indicated in Table 1 and Tables S1, S2, S3 (supplementary material).
. All utilized parameters are indicated in Table 1 and Tables S1, S2, S3 (supplementary material).
 cells. The light grey area indicates the in vivo concentrations range of TFV-DP from , , , converted to units
cells. The light grey area indicates the in vivo concentrations range of TFV-DP from , , , converted to units  by assuming a cell volume of
by assuming a cell volume of  for resting
for resting  T-cells . The dashed horizontal line indicates the threshold for resistance selection, i.e.
T-cells . The dashed horizontal line indicates the threshold for resistance selection, i.e.  . Panel A: Selective advantage of the respective mutants with regard to wild type
. Panel A: Selective advantage of the respective mutants with regard to wild type  . B: Selective advantage of a succeeding mutants with regard to progenitor in Q151M complex formation
. B: Selective advantage of a succeeding mutants with regard to progenitor in Q151M complex formation  . All utilized parameters are indicated in Table 1 and Tables S1, S2 (supplementary material).
. All utilized parameters are indicated in Table 1 and Tables S1, S2 (supplementary material).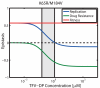
 , resistance
, resistance  and fitness
and fitness  as defined in eqs. (5)–(7) for the double mutant ‘K65R/M184V’. The black dashed horizontal line indicates the value, where no epistatic interactions occur. The light grey area indicates the in vivo concentrations range of TFV-DP from , , , converted to units
as defined in eqs. (5)–(7) for the double mutant ‘K65R/M184V’. The black dashed horizontal line indicates the value, where no epistatic interactions occur. The light grey area indicates the in vivo concentrations range of TFV-DP from , , , converted to units  by assuming a cell volume of
by assuming a cell volume of  for unstimulated
for unstimulated  T-cells . All utilized parameters are indicated in Table 1 and Tables S1, S2 (supplementary material).
T-cells . All utilized parameters are indicated in Table 1 and Tables S1, S2 (supplementary material).Similar articles
-
Molecular mechanism by which the K70E mutation in human immunodeficiency virus type 1 reverse transcriptase confers resistance to nucleoside reverse transcriptase inhibitors.Antimicrob Agents Chemother. 2007 Jan;51(1):48-53. doi: 10.1128/AAC.00683-06. Epub 2006 Nov 6. Antimicrob Agents Chemother. 2007. PMID: 17088490 Free PMC article.
-
Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision.J Virol. 2002 Sep;76(18):9143-51. doi: 10.1128/jvi.76.18.9143-9151.2002. J Virol. 2002. PMID: 12186898 Free PMC article.
-
The L74V mutation in human immunodeficiency virus type 1 reverse transcriptase counteracts enhanced excision of zidovudine monophosphate associated with thymidine analog resistance mutations.Antimicrob Agents Chemother. 2005 Jul;49(7):2648-56. doi: 10.1128/AAC.49.7.2648-2656.2005. Antimicrob Agents Chemother. 2005. PMID: 15980332 Free PMC article.
-
Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure.Antivir Ther. 2001;6 Suppl 3:25-44. Antivir Ther. 2001. PMID: 11678471 Review.
-
Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs).Cell Mol Life Sci. 2000 Sep;57(10):1408-22. doi: 10.1007/PL00000626. Cell Mol Life Sci. 2000. PMID: 11078020 Free PMC article. Review.
Cited by
-
In vitro HIV-1 evolution in response to triple reverse transcriptase inhibitors & in silico phenotypic analysis.PLoS One. 2013 Apr 17;8(4):e61102. doi: 10.1371/journal.pone.0061102. Print 2013. PLoS One. 2013. PMID: 23613794 Free PMC article.
-
Modeling of HIV-1 prophylactic efficacy and toxicity with islatravir shows non-superiority for oral dosing, but promise as a subcutaneous implant.CPT Pharmacometrics Syst Pharmacol. 2024 Oct;13(10):1693-1706. doi: 10.1002/psp4.13212. Epub 2024 Aug 20. CPT Pharmacometrics Syst Pharmacol. 2024. PMID: 39164932 Free PMC article.
-
The Restrictome of Flaviviruses.Virol Sin. 2020 Aug;35(4):363-377. doi: 10.1007/s12250-020-00208-3. Epub 2020 Mar 9. Virol Sin. 2020. PMID: 32152893 Free PMC article. Review.
-
Mechanistic framework predicts drug-class specific utility of antiretrovirals for HIV prophylaxis.PLoS Comput Biol. 2019 Jan 30;15(1):e1006740. doi: 10.1371/journal.pcbi.1006740. eCollection 2019 Jan. PLoS Comput Biol. 2019. PMID: 30699105 Free PMC article.
-
Estimating HIV-1 fitness characteristics from cross-sectional genotype data.PLoS Comput Biol. 2014 Nov 6;10(11):e1003886. doi: 10.1371/journal.pcbi.1003886. eCollection 2014 Nov. PLoS Comput Biol. 2014. PMID: 25375675 Free PMC article.
References
-
- Clercq ED. Strategies in the design of antiviral drugs. Nat Rev Drug Discov. 2002;1:925 13–25. - PubMed
-
- Tsai CH, Lee PY, Stollar V, Li ML. Antiviral therapy targeting viral poly-merase. Curr Pharm Des. 2006;12:1339–1355. - PubMed
-
- Straus SE, Takiff HE, Seidlin M, Bachrach S, Lininger L, et al. Suppression of frequently recurring genital herpes. A placebo-controlled double-blind trial of oral acyclovir. N Engl J Med. 1984;310:1545–1550. - PubMed
-
- Douglas JM, Critchlow C, Benedetti J, Mertz GJ, Connor JD, et al. A double blind study of oral acyclovir for suppression of recurrences of genital herpes simplex 3 virus infection. N Engl J Med. 1984;310:1551–1556. - PubMed
-
- Fischl MA, Richman DD, Grieco MH, Gottlieb MS, Volberding PA, et al. The efficacy of azidothymidine (AZT) in the treatment of patients with AIDS and AIDS-related complex. A double-blind, placebo-controlled trial. N Engl J Med. 1987;317:185–191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

