Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression
- PMID: 22275870
- PMCID: PMC3261927
- DOI: 10.1371/journal.pgen.1002431
Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression
Abstract
It is known that genetic variants can affect gene expression, but it is not yet completely clear through what mechanisms genetic variation mediate this expression. We therefore compared the cis-effect of single nucleotide polymorphisms (SNPs) on gene expression between blood samples from 1,240 human subjects and four primary non-blood tissues (liver, subcutaneous, and visceral adipose tissue and skeletal muscle) from 85 subjects. We characterized four different mechanisms for 2,072 probes that show tissue-dependent genetic regulation between blood and non-blood tissues: on average 33.2% only showed cis-regulation in non-blood tissues; 14.5% of the eQTL probes were regulated by different, independent SNPs depending on the tissue of investigation. 47.9% showed a different effect size although they were regulated by the same SNPs. Surprisingly, we observed that 4.4% were regulated by the same SNP but with opposite allelic direction. We show here that SNPs that are located in transcriptional regulatory elements are enriched for tissue-dependent regulation, including SNPs at 3' and 5' untranslated regions (P = 1.84×10(-5) and 4.7×10(-4), respectively) and SNPs that are synonymous-coding (P = 9.9×10(-4)). SNPs that are associated with complex traits more often exert a tissue-dependent effect on gene expression (P = 2.6×10(-10)). Our study yields new insights into the genetic basis of tissue-dependent expression and suggests that complex trait associated genetic variants have even more complex regulatory effects than previously anticipated.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
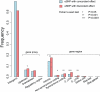
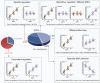
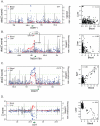

References
-
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. - DOI - PMC - PubMed
-
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen K, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

