Cohesin protects genes against γH2AX Induced by DNA double-strand breaks
- PMID: 22275873
- PMCID: PMC3261922
- DOI: 10.1371/journal.pgen.1002460
Cohesin protects genes against γH2AX Induced by DNA double-strand breaks
Abstract
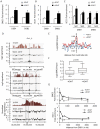
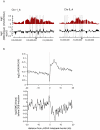
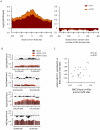
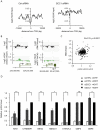
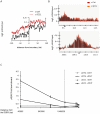
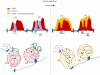
Chromatin undergoes major remodeling around DNA double-strand breaks (DSB) to promote repair and DNA damage response (DDR) activation. We recently reported a high-resolution map of γH2AX around multiple breaks on the human genome, using a new cell-based DSB inducible system. In an attempt to further characterize the chromatin landscape induced around DSBs, we now report the profile of SMC3, a subunit of the cohesin complex, previously characterized as required for repair by homologous recombination. We found that recruitment of cohesin is moderate and restricted to the immediate vicinity of DSBs in human cells. In addition, we show that cohesin controls γH2AX distribution within domains. Indeed, as we reported previously for transcription, cohesin binding antagonizes γH2AX spreading. Remarkably, depletion of cohesin leads to an increase of γH2AX at cohesin-bound genes, associated with a decrease in their expression level after DSB induction. We propose that, in agreement with their function in chromosome architecture, cohesin could also help to isolate active genes from some chromatin remodelling and modifications such as the ones that occur when a DSB is detected on the genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
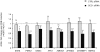
References
-
- van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6:757–765. - PubMed
-
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. - PubMed
-
- Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J Biol Chem. 2003;278:19579–19582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

