Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression
- PMID: 22276136
- PMCID: PMC3262799
- DOI: 10.1371/journal.pone.0029979
Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression
Abstract
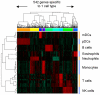
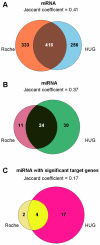
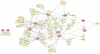
Blood consists of different cell populations with distinct functions and correspondingly, distinct gene expression profiles. In this study, global miRNA expression profiling was performed across a panel of nine human immune cell subsets (neutrophils, eosinophils, monocytes, B cells, NK cells, CD4 T cells, CD8 T cells, mDCs and pDCs) to identify cell-type specific miRNAs. mRNA expression profiling was performed on the same samples to determine if miRNAs specific to certain cell types down-regulated expression levels of their target genes. Six cell-type specific miRNAs (miR-143; neutrophil specific, miR-125; T cells and neutrophil specific, miR-500; monocyte and pDC specific, miR-150; lymphoid cell specific, miR-652 and miR-223; both myeloid cell specific) were negatively correlated with expression of their predicted target genes. These results were further validated using an independent cohort where similar immune cell subsets were isolated and profiled for both miRNA and mRNA expression. miRNAs which negatively correlated with target gene expression in both cohorts were identified as candidates for miRNA/mRNA regulatory pairs and were used to construct a cell-type specific regulatory network. miRNA/mRNA pairs formed two distinct clusters in the network corresponding to myeloid (nine miRNAs) and lymphoid lineages (two miRNAs). Several myeloid specific miRNAs targeted common genes including ABL2, EIF4A2, EPC1 and INO80D; these common targets were enriched for genes involved in the regulation of gene expression (p<9.0E-7). Those miRNA might therefore have significant further effect on gene expression by repressing the expression of genes involved in transcriptional regulation. The miRNA and mRNA expression profiles reported in this study form a comprehensive transcriptome database of various human blood cells and serve as a valuable resource for elucidating the role of miRNA mediated regulation in the establishment of immune cell identity.
Conflict of interest statement
Figures


References
-
- Lindstedt M, Lundberg K, Borrebaeck CA. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J Immunol. 2005;175:4839–4846. - PubMed
-
- Zhao C, Zhang H, Wong WC, Sem X, Han H, et al. Identification of novel functional differences in monocyte subsets using proteomic and transcriptomic methods. J Proteome Res. 2009;8:4028–4038. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

