Bacterial community assembly and turnover within the intestines of developing zebrafish
- PMID: 22276219
- PMCID: PMC3261916
- DOI: 10.1371/journal.pone.0030603
Bacterial community assembly and turnover within the intestines of developing zebrafish
Abstract
Background: The majority of animal associated microorganisms are present in digestive tract communities. These intestinal communities arise from selective pressures of the gut habitats as well as host's genotype are regarded as an extra 'organ' regulate functions that have not evolved wholly on the host. They are functionally essential in providing nourishment, regulating epithelial development, and influencing immunity in the vertebrate host. As vertebrates are born free of microorganisms, what is poorly understood is how intestinal bacterial communities assemble and develop in conjunction with the development of the host.
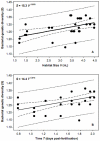
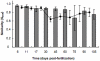
Methodology/principal findings: Set within an ecological framework, we investigated the bacterial community assembly and turnover within the intestinal habitats of developing zebrafish (from larvae to adult animals). Spatial and temporal species-richness relationships and Mantel and partial Mantel tests revealed that turnover was low and that richness and composition was best predicted by time and not intestinal volume (habitat size) or changes in food diet. We also observed that bacterial communities within the zebrafish intestines were deterministically assembled (reflected by the observed low turnover) switching to stochastic assembly in the later stages of zebrafish development.
Conclusions/significance: This study is of importance as it provides a novel insight into how intestinal bacterial communities assemble in tandem with the host's development (from early to adult stages). It is our hope that by studying intestinal microbiota of this vertebrate model with such or some more refined approaches in the future could well provide ecological insights for clinical benefit. In addition, this study also adds to our still fledgling knowledge of how spatial and temporal species-richness relationships are shaped and provides further mounting evidence that bacterial community assembly and dynamics are shaped by both deterministic and stochastic considerations.
Conflict of interest statement
Figures

References
-
- Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, et al. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–386. - PubMed
-
- Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–1051S. - PubMed
-
- Rawls JF. Microbial community assembly and host-microbial interactions in the intestine. Pediatr Pulmonol. 2007;S30:146–148.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

