Protein-DNA interactions define the mechanistic aspects of circle formation and insertion reactions in IS2 transposition
- PMID: 22277150
- PMCID: PMC3299598
- DOI: 10.1186/1759-8753-3-1
Protein-DNA interactions define the mechanistic aspects of circle formation and insertion reactions in IS2 transposition
Abstract
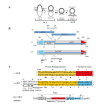
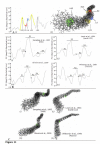
Background: Transposition in IS3, IS30, IS21 and IS256 insertion sequence (IS) families utilizes an unconventional two-step pathway. A figure-of-eight intermediate in Step I, from asymmetric single-strand cleavage and joining reactions, is converted into a double-stranded minicircle whose junction (the abutted left and right ends) is the substrate for symmetrical transesterification attacks on target DNA in Step II, suggesting intrinsically different synaptic complexes (SC) for each step. Transposases of these ISs bind poorly to cognate DNA and comparative biophysical analyses of SC I and SC II have proven elusive. We have prepared a native, soluble, active, GFP-tagged fusion derivative of the IS2 transposase that creates fully formed complexes with single-end and minicircle junction (MCJ) substrates and used these successfully in hydroxyl radical footprinting experiments.
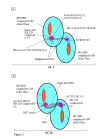
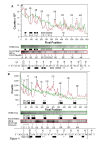
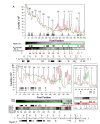
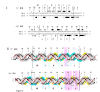
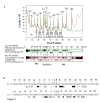
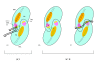
Results: In IS2, Step I reactions are physically and chemically asymmetric; the left imperfect, inverted repeat (IRL), the exclusive recipient end, lacks donor function. In SC I, different protection patterns of the cleavage domains (CDs) of the right imperfect inverted repeat (IRR; extensive in cis) and IRL (selective in trans) at the single active cognate IRR catalytic center (CC) are related to their donor and recipient functions. In SC II, extensive binding of the IRL CD in trans and of the abutted IRR CD in cis at this CC represents the first phase of the complex. An MCJ substrate precleaved at the 3' end of IRR revealed a temporary transition state with the IRL CD disengaged from the protein. We propose that in SC II, sequential 3' cleavages at the bound abutted CDs trigger a conformational change, allowing the IRL CD to complex to its cognate CC, producing the second phase. Corroborating data from enhanced residues and curvature propensity plots suggest that CD to CD interactions in SC I and SC II require IRL to assume a bent structure, to facilitate binding in trans.
Conclusions: Different transpososomes are assembled in each step of the IS2 transposition pathway. Recipient versus donor end functions of the IRL CD in SC I and SC II and the conformational change in SC II that produces the phase needed for symmetrical IRL and IRR donor attacks on target DNA highlight the differences.
Figures



References
-
- Chandler M, Mahillon J. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editor. Washington, DC: ASM Press; 2002. Insertion sequences revisited; pp. 305–366.
-
- Rousseau P, Normand C, Loot C, Turlan C, Alazard R, Duval-Valentin G, Chandler M. In: Mobile DNA I. Craig NL, Craigie R, Gellert M, Lambowitz AM, editor. Washington, DC: ASM Press; 2002. Transposition of IS911; pp. 367–383.
LinkOut - more resources
Full Text Sources

