High-throughput decoding of antitrypanosomal drug efficacy and resistance
- PMID: 22278056
- PMCID: PMC3303116
- DOI: 10.1038/nature10771
High-throughput decoding of antitrypanosomal drug efficacy and resistance
Abstract
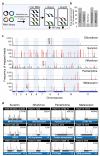
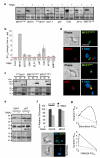
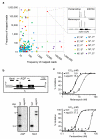
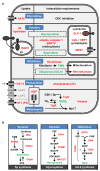
The concept of disease-specific chemotherapy was developed a century ago. Dyes and arsenical compounds that displayed selectivity against trypanosomes were central to this work, and the drugs that emerged remain in use for treating human African trypanosomiasis (HAT). The importance of understanding the mechanisms underlying selective drug action and resistance for the development of improved HAT therapies has been recognized, but these mechanisms have remained largely unknown. Here we use all five current HAT drugs for genome-scale RNA interference target sequencing (RIT-seq) screens in Trypanosoma brucei, revealing the transporters, organelles, enzymes and metabolic pathways that function to facilitate antitrypanosomal drug action. RIT-seq profiling identifies both known drug importers and the only known pro-drug activator, and links more than fifty additional genes to drug action. A bloodstream stage-specific invariant surface glycoprotein (ISG75) family mediates suramin uptake, and the AP1 adaptin complex, lysosomal proteases and major lysosomal transmembrane protein, as well as spermidine and N-acetylglucosamine biosynthesis, all contribute to suramin action. Further screens link ubiquinone availability to nitro-drug action, plasma membrane P-type H(+)-ATPases to pentamidine action, and trypanothione and several putative kinases to melarsoprol action. We also demonstrate a major role for aquaglyceroporins in pentamidine and melarsoprol cross-resistance. These advances in our understanding of mechanisms of antitrypanosomal drug efficacy and resistance will aid the rational design of new therapies and help to combat drug resistance, and provide unprecedented molecular insight into the mode of action of antitrypanosomal drugs.
Figures
Comment in
-
Infectious disease: Genomics decodes drug action.Nature. 2012 Feb 8;482(7384):167-9. doi: 10.1038/482167a. Nature. 2012. PMID: 22318598 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

