Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist
- PMID: 22278061
- PMCID: PMC3345277
- DOI: 10.1038/nature10753
Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist
Abstract
The parasympathetic branch of the autonomic nervous system regulates the activity of multiple organ systems. Muscarinic receptors are G-protein-coupled receptors that mediate the response to acetylcholine released from parasympathetic nerves. Their role in the unconscious regulation of organ and central nervous system function makes them potential therapeutic targets for a broad spectrum of diseases. The M2 muscarinic acetylcholine receptor (M2 receptor) is essential for the physiological control of cardiovascular function through activation of G-protein-coupled inwardly rectifying potassium channels, and is of particular interest because of its extensive pharmacological characterization with both orthosteric and allosteric ligands. Here we report the structure of the antagonist-bound human M2 receptor, the first human acetylcholine receptor to be characterized structurally, to our knowledge. The antagonist 3-quinuclidinyl-benzilate binds in the middle of a long aqueous channel extending approximately two-thirds through the membrane. The orthosteric binding pocket is formed by amino acids that are identical in all five muscarinic receptor subtypes, and shares structural homology with other functionally unrelated acetylcholine binding proteins from different species. A layer of tyrosine residues forms an aromatic cap restricting dissociation of the bound ligand. A binding site for allosteric ligands has been mapped to residues at the entrance to the binding pocket near this aromatic cap. The structure of the M2 receptor provides insights into the challenges of developing subtype-selective ligands for muscarinic receptors and their propensity for allosteric regulation.
Figures


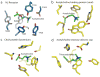
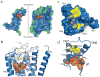
Comment in
-
Structural biology: Muscarinic receptors become crystal clear.Nature. 2012 Feb 22;482(7386):480-1. doi: 10.1038/482480a. Nature. 2012. PMID: 22358836 No abstract available.
References
-
- Hulme EC, Birdsall NJ, Buckley NJ. Muscarinic receptor subtypes. Annu Rev Pharmacol and Toxicol. 1990;30:633–673. - PubMed
-
- Dale HH. The action of certain esters and ethers of choline, and their relation to muscarine. J Pharmacol Exp Ther. 1914;6:147–190.
-
- Haga K, et al. Functional reconstitution of purified muscarinic receptors and inhibitory guanine nucleotide regulatory protein. Nature. 1985;316:731–733. - PubMed
-
- Haga K, Haga T. Purification of the muscarinic acetylcholine receptor from porcine brain. J Biol Chem. 1985;260:7927–7935. - PubMed
-
- Kubo T, et al. Cloning, sequencing and expression of complementary DNA encoding the muscarinic acetylcholine receptor. Nature. 1986;323:411–416. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

