The conserved 5' apical hairpin stem loops of bamboo mosaic virus and its satellite RNA contribute to replication competence
- PMID: 22278884
- PMCID: PMC3378871
- DOI: 10.1093/nar/gks030
The conserved 5' apical hairpin stem loops of bamboo mosaic virus and its satellite RNA contribute to replication competence
Abstract
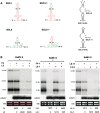
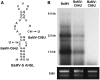
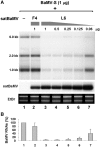
Satellite RNAs associated with Bamboo mosaic virus (satBaMVs) depend on BaMV for replication and encapsidation. Certain satBaMVs isolated from natural fields significantly interfere with BaMV replication. The 5' apical hairpin stem loop (AHSL) of satBaMV is the major determinant in interference with BaMV replication. In this study, by in vivo competition assay, we revealed that the sequence and structure of AHSL, along with specific nucleotides (C(60) and C(83)) required for interference with BaMV replication, are also involved in replication competition among satBaMV variants. Moreover, all of the 5' ends of natural BaMV isolates contain the similar AHSLs having conserved nucleotides (C(64) and C(86)) with those of interfering satBaMVs, suggesting their co-evolution. Mutational analyses revealed that C(86) was essential for BaMV replication, and that replacement of C(64) with U reduced replication efficiency. The non-interfering satBaMV interfered with BaMV replication with the BaMV-C64U mutant as helper. These findings suggest that two cytosines at the equivalent positions in the AHSLs of BaMV and satBaMV play a crucial role in replication competence. The downregulation level, which is dependent upon the molar ratio of interfering satBaMV to BaMV, implies that there is competition for limited replication machinery.
Figures




Similar articles
-
Interfering Satellite RNAs of Bamboo mosaic virus.Front Microbiol. 2017 May 4;8:787. doi: 10.3389/fmicb.2017.00787. eCollection 2017. Front Microbiol. 2017. PMID: 28522996 Free PMC article. Review.
-
Crucial role of the 5' conserved structure of bamboo mosaic virus satellite RNA in downregulation of helper viral RNA replication.J Virol. 2006 Mar;80(5):2566-74. doi: 10.1128/JVI.80.5.2566-2574.2006. J Virol. 2006. PMID: 16474162 Free PMC article.
-
Downregulation of Bamboo mosaic virus replication requires the 5' apical hairpin stem loop structure and sequence of satellite RNA.Virology. 2007 Sep 1;365(2):271-84. doi: 10.1016/j.virol.2007.03.050. Epub 2007 May 4. Virology. 2007. PMID: 17482233
-
Structural and functional analyses of the 3' untranslated region of Bamboo mosaic virus satellite RNA.Virology. 2009 Mar 30;386(1):139-53. doi: 10.1016/j.virol.2009.01.019. Epub 2009 Feb 7. Virology. 2009. PMID: 19201437
-
Viral elements and host cellular proteins in intercellular movement of Bamboo mosaic virus.Curr Opin Virol. 2015 Jun;12:99-108. doi: 10.1016/j.coviro.2015.04.005. Epub 2015 May 13. Curr Opin Virol. 2015. PMID: 25951346 Review.
Cited by
-
Interfering Satellite RNAs of Bamboo mosaic virus.Front Microbiol. 2017 May 4;8:787. doi: 10.3389/fmicb.2017.00787. eCollection 2017. Front Microbiol. 2017. PMID: 28522996 Free PMC article. Review.
-
Two key arginine residues in the coat protein of Bamboo mosaic virus differentially affect the accumulation of viral genomic and subgenomic RNAs.Mol Plant Pathol. 2014 Feb;15(2):196-210. doi: 10.1111/mpp.12080. Epub 2013 Oct 28. Mol Plant Pathol. 2014. PMID: 24393453 Free PMC article.
-
Genetic diversity and evolution of satellite RNAs associated with the bamboo mosaic virus.PLoS One. 2014 Oct 2;9(9):e108015. doi: 10.1371/journal.pone.0108015. eCollection 2014. PLoS One. 2014. PMID: 25275532 Free PMC article.
-
Trade-off between local replication and long-distance dissemination during experimental evolution of a satellite RNA.Front Microbiol. 2023 Aug 4;14:1139447. doi: 10.3389/fmicb.2023.1139447. eCollection 2023. Front Microbiol. 2023. PMID: 37601360 Free PMC article.
-
Biologically-supported structural model for a viral satellite RNA.Nucleic Acids Res. 2015 Nov 16;43(20):9965-77. doi: 10.1093/nar/gkv917. Epub 2015 Sep 17. Nucleic Acids Res. 2015. PMID: 26384416 Free PMC article.
References
-
- Mandahar CL. Multiplication of RNA plant viruses. Springer. 2006:10–14.
-
- Lough TJ, Lee RH, Emerson SJ, Forster RL, Lucas WJ. Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology. 2006;351:455–465. - PubMed

