Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize
- PMID: 22279089
- PMCID: PMC3325079
- DOI: 10.1093/dnares/dsr048
Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize
Abstract
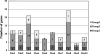
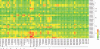
The WRKY transcription factors function in plant growth and development, and response to the biotic and abiotic stresses. Although many studies have focused on the functional identification of the WRKY transcription factors, much less is known about molecular phylogenetic and global expression analysis of the complete WRKY family in maize. In this study, we identified 136 WRKY proteins coded by 119 genes in the B73 inbred line from the complete genome and named them in an orderly manner. Then, a comprehensive phylogenetic analysis of five species was performed to explore the origin and evolutionary patterns of these WRKY genes, and the result showed that gene duplication is the major driving force for the origin of new groups and subgroups and functional divergence during evolution. Chromosomal location analysis of maize WRKY genes indicated that 20 gene clusters are distributed unevenly in the genome. Microarray-based expression analysis has revealed that 131 WRKY transcripts encoded by 116 genes may participate in the regulation of maize growth and development. Among them, 102 transcripts are stably expressed with a coefficient of variation (CV) value of <15%. The remaining 29 transcripts produced by 25 WRKY genes with the CV value of >15% are further analysed to discover new organ- or tissue-specific genes. In addition, microarray analyses of transcriptional responses to drought stress and fungal infection showed that maize WRKY proteins are involved in stress responses. All these results contribute to a deep probing into the roles of WRKY transcription factors in maize growth and development and stress tolerance.
Figures



References
-
- Ulker B., Somssich I.E. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

