Protein design using continuous rotamers
- PMID: 22279426
- PMCID: PMC3257257
- DOI: 10.1371/journal.pcbi.1002335
Protein design using continuous rotamers
Abstract
Optimizing amino acid conformation and identity is a central problem in computational protein design. Protein design algorithms must allow realistic protein flexibility to occur during this optimization, or they may fail to find the best sequence with the lowest energy. Most design algorithms implement side-chain flexibility by allowing the side chains to move between a small set of discrete, low-energy states, which we call rigid rotamers. In this work we show that allowing continuous side-chain flexibility (which we call continuous rotamers) greatly improves protein flexibility modeling. We present a large-scale study that compares the sequences and best energy conformations in 69 protein-core redesigns using a rigid-rotamer model versus a continuous-rotamer model. We show that in nearly all of our redesigns the sequence found by the continuous-rotamer model is different and has a lower energy than the one found by the rigid-rotamer model. Moreover, the sequences found by the continuous-rotamer model are more similar to the native sequences. We then show that the seemingly easy solution of sampling more rigid rotamers within the continuous region is not a practical alternative to a continuous-rotamer model: at computationally feasible resolutions, using more rigid rotamers was never better than a continuous-rotamer model and almost always resulted in higher energies. Finally, we present a new protein design algorithm based on the dead-end elimination (DEE) algorithm, which we call iMinDEE, that makes the use of continuous rotamers feasible in larger systems. iMinDEE guarantees finding the optimal answer while pruning the search space with close to the same efficiency of DEE.
Availability: Software is available under the Lesser GNU Public License v3. Contact the authors for source code.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
 and
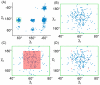
and  angles) and the ocurrence of isoleucine conformations across a wide set of high-quality structures is plotted here. Panel A shows the entire
angles) and the ocurrence of isoleucine conformations across a wide set of high-quality structures is plotted here. Panel A shows the entire  and
and  angle space, while panels B, C, and D zoom in on the region specific to one rotamer. (A) The side chains of amino acids commonly appear almost exclusively (blue dots in the plot) within specific regions of their flexible space. (B) In a rigid-rotamer model a single conformation (the red diamond) represents that entire region. (C) In a continuous-rotamer model, a voxel models the continuous region that represents the rotamer. (D) An expanded rotamer model samples additional rigid rotamers near rotamers from the rigid-rotamer model.
angle space, while panels B, C, and D zoom in on the region specific to one rotamer. (A) The side chains of amino acids commonly appear almost exclusively (blue dots in the plot) within specific regions of their flexible space. (B) In a rigid-rotamer model a single conformation (the red diamond) represents that entire region. (C) In a continuous-rotamer model, a voxel models the continuous region that represents the rotamer. (D) An expanded rotamer model samples additional rigid rotamers near rotamers from the rigid-rotamer model.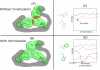
 -angle space can have profound effects on the energies of interacting rotamers, particularly in the packed core of a protein. The three hydrophobic residues in this toy example can form a well-packed core through small changes in their
-angle space can have profound effects on the energies of interacting rotamers, particularly in the packed core of a protein. The three hydrophobic residues in this toy example can form a well-packed core through small changes in their  angles in this cartoon. A pruning algorithm like rigid DEE would erroneously prune the clashing rotamers since it does not account for these small changes. (C) If one rotamer,
angles in this cartoon. A pruning algorithm like rigid DEE would erroneously prune the clashing rotamers since it does not account for these small changes. (C) If one rotamer,  , always results in conformations of higher energy than another,
, always results in conformations of higher energy than another,  , the rotamer
, the rotamer  and all the conformations that contain
and all the conformations that contain  can be pruned. The rigid DEE algorithm prunes rotamers and amino acids that are provably not part of the rigid GMEC. (D) When rotamers can minimize within their specified voxel, rotamers and amino acids that seemed poor in a rigid model might minimize to lower energy conformations than the rigid GMEC. The lowest-energy conformation in this scenario is the minGMEC. The MinDEE algorithm and iMinDEE algorithm can provably prune rotamers in the presence of minimization.
can be pruned. The rigid DEE algorithm prunes rotamers and amino acids that are provably not part of the rigid GMEC. (D) When rotamers can minimize within their specified voxel, rotamers and amino acids that seemed poor in a rigid model might minimize to lower energy conformations than the rigid GMEC. The lowest-energy conformation in this scenario is the minGMEC. The MinDEE algorithm and iMinDEE algorithm can provably prune rotamers in the presence of minimization.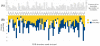

 ) has been found, but unpruned high energy conformations slow down the search. The first conformation enumerated by A*, corresponding to the conformation with the lowest energy bound, is denoted
) has been found, but unpruned high energy conformations slow down the search. The first conformation enumerated by A*, corresponding to the conformation with the lowest energy bound, is denoted  , and the lower bound on its energy is
, and the lower bound on its energy is  . The minGMEC,
. The minGMEC,  , is marked by a green dot and its energy is
, is marked by a green dot and its energy is  . (B) Instead of MinDEE, we can use iMinDEE to prune conformations with energy bounds that are higher than the lowest energy bound by more than the initial
. (B) Instead of MinDEE, we can use iMinDEE to prune conformations with energy bounds that are higher than the lowest energy bound by more than the initial  value. We then select the lowest minimized energy found so far (i.e. as opposed to lowest energy bound) and use that to compute the
value. We then select the lowest minimized energy found so far (i.e. as opposed to lowest energy bound) and use that to compute the  value. The conformation with the lowest minimized energy is denoted
value. The conformation with the lowest minimized energy is denoted  with a blue dot and its energy is
with a blue dot and its energy is  . (C) The iMinDEE search is repeated if
. (C) The iMinDEE search is repeated if  . Since
. Since  ,
,  meets the condition of Eq. (6), and the search will not need to be repeated again. By setting
meets the condition of Eq. (6), and the search will not need to be repeated again. By setting  , we can use the iMinDEE criterion (Eq. (7)) to prune rotamers, and the iMinDEE algorithm will provably find the minGMEC.
, we can use the iMinDEE criterion (Eq. (7)) to prune rotamers, and the iMinDEE algorithm will provably find the minGMEC.

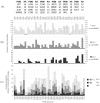
 values. Some outliers have larger
values. Some outliers have larger  values, and in consequence, iMinDEE loses pruning efficiency in these systems.
values, and in consequence, iMinDEE loses pruning efficiency in these systems.

References
-
- Gorczynski MJ, Grembecka J, Zhou Y, Kong Y, Roudaia L, et al. Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBFβ. Chem Biol. 2007;14:1186–97. - PubMed
-
- Roberts KE, Cushing PR, Boisguerin P, Madden DR, Donald BR. Research in Com- putational Molecular Biology. volume 6577 of Lecture Notes in Computer Science. Heidelberg: Springer Berlin; 2011. Design of protein- protein interactions with a novel ensemble-based scoring algorithm. pp. 361–376.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

